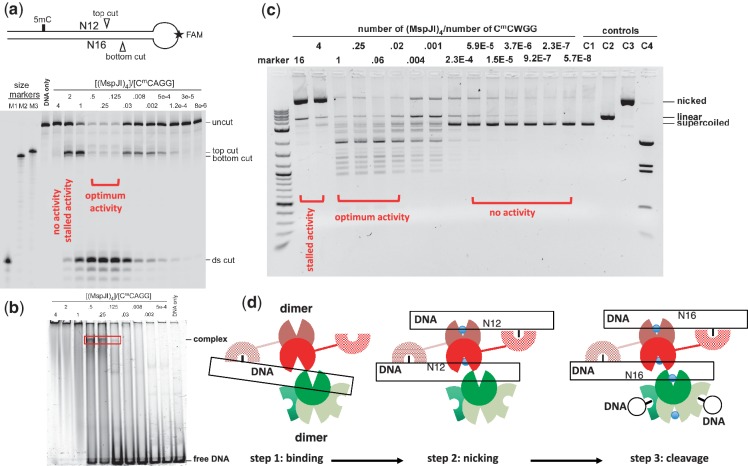
Figure 6.
MspJI cleaves via a top-strand nicked intermediate. (a) A stem-loop structured oligonucleotide substrate containing one 5mC site was designed with an internal fluorescent FAM label in the loop. Size markers were synthesized according to predicted cleavage sites: M1, product from a ds cleavage; M2, product from the bottom-strand cleavage; M3, product from the top-strand cleavage. A 2-fold titration of MspJI digestion started from the tetramer to DNA ratio of 4 to 0.125 and followed by 4-fold dilution. (b) DNA-binding assays were performed by incubating 0.5 µM FAM labeled stem-loop oligonucleotides with varying amount of MspJI tetramer at 37°C for 1 h. (c) MspJI titration on pBR322 (dcm+) containing six C5mCWGG sites. The molar ratio of MspJI tetramer to its substrate sites is shown on the top of the lanes. Control lanes include: C1, pBR322 only; C2, pBR322 digested with EcoRI, which produces linearized pBR322; C3, pBR322 digested with nicking endonuclease Nt.BspQI, which produces a nicked pBR322; C4, pBR322 digested with BstNI, which produces a complete digestion pattern at C5mCWGG. (d) A proposed three-step mechanism of the MspJI enzymatic reaction. Step 1: one SRA-like domain binds specifically to the modified cytosine. With a tetramer-to-DNA ratio of 4:1, no enzymatic activity was observed. Step 2: the other SRA-like domain of the same back-to-back dimer binds another target site, resulting in a top-strand nicked intermediate. With a tetramer-to-DNA ratio of 2:1, the reaction stalled after the first N12 cut. Step 3: with a tetramer-to-DNA ratio of 1:4, the highest level of ds cleavage was observed.