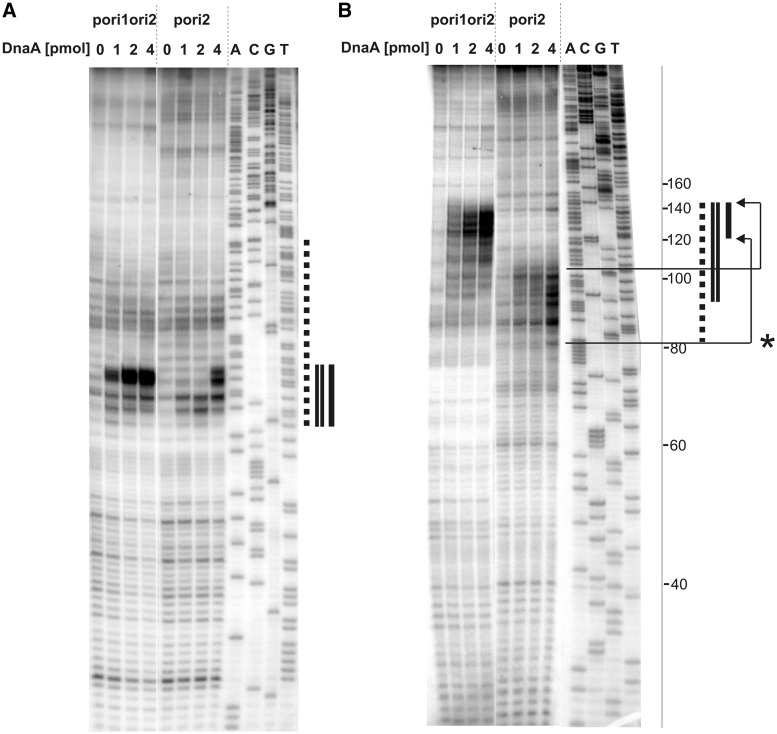
Figure 3.
Determination of the H. pylori oriC sequence unwound by DnaA in vitro. Plasmid DNA, after incubation with the indicated amounts of the DnaA protein and P1 nuclease treatment, was used as a substrate for PE analysis. 32P labeled primers P-9 and P-10 were complementary to the coding strand (with respect to the dnaA gene) (A) and non-coding strand (B), respectively. Dotted line corresponds to the AT-rich region identified in silico as a DUE. Single and double lines refer to the areas susceptible to P1 nuclease digestion in pori2 and pori1ori2 plasmids, respectively. A, C, G, T sequencing reactions were carried out with 32P labeled primers P-9 (A), P-10 (B) and the pori1ori2 plasmid DNA. The ladder on the right side of the figure corresponds to the distance from P-10 primer annealing site on pori1ori2. The P-10 PE product on pori2 is 40 bps shorter than that on pori1ori2 (Supplementary Figure S4). Thus to determine the position of the unwound region on pori2 it is necessary to add 40 bp to the observed PE bands positions; * - arrows correspond to the actual position of the pori2 unwound region within oriC2.