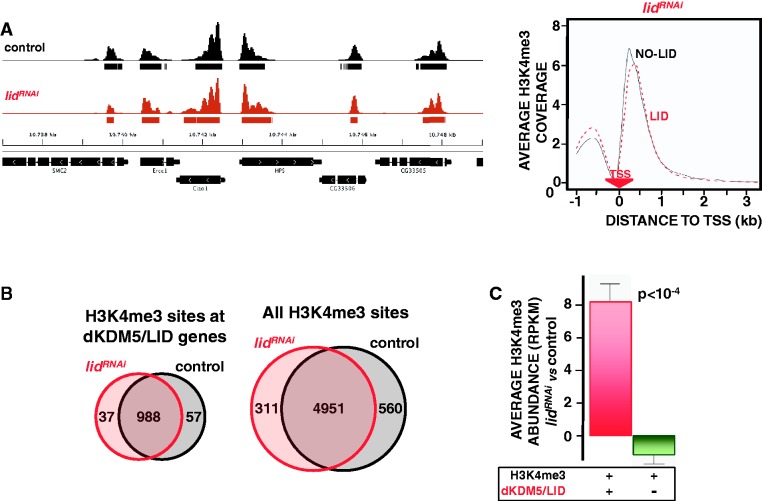
Figure 3.
dKDM5/LID depletion increases H3K4me3 at TSS. (A) ChIP-seq coverage profiles of H3K4me3 across a representative region are presented both in control wt flies (black) and lidRNAi knockdown flies (red), where dKDM5/LID depletion was induced by the Actin5C-GAL4 driver in heterozygous lidk06801/+ mutant flies. Rectangles underneath each profile indicate the position of the corresponding peaks/binding sites. Genomic organization of the region is indicated. On the right, H3K4me3 distribution around TSS in lidRNAi knockdown flies is presented for genes containing dKDM5/LID (red) or not (black). For each gene, the coverage profile was normalized dividing by the average coverage in that gene. The position of the TSS is indicated. (B) Venn diagrams showing the intersection between H3K4me3 sites in control wt (black) and lidRNAi knockdown flies (red) detected at dKDM5/LID target genes (left) and when all H3K4me3 sites are considered (right). (C) Relative H3K4me3 abundance in lidRNAi knockdown flies versus control wt flies is presented for genes containing both dKDM5/LID and H3K4me3 (red) and genes containing only H3K4me3 (green). Statistical significance of the difference (Kruskal–Wallis P-value) is indicated.