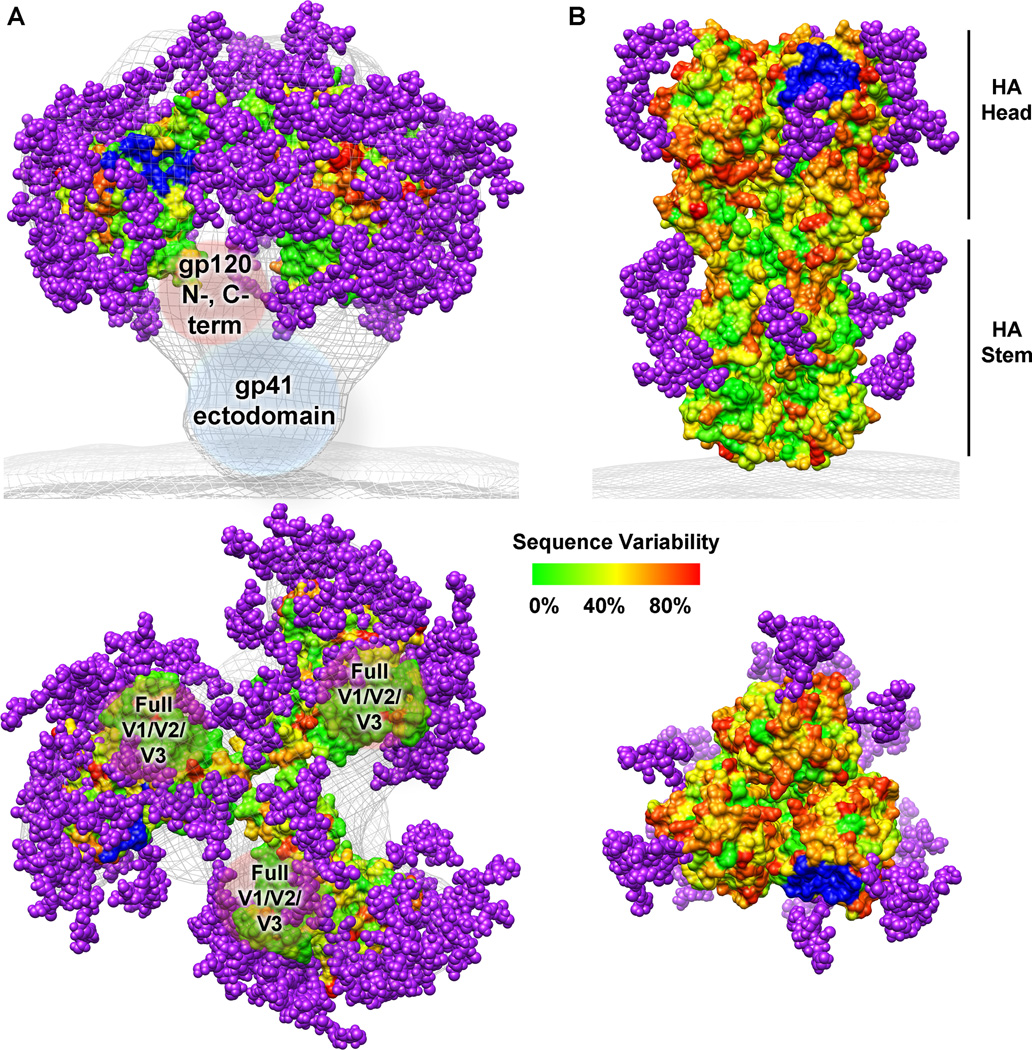
Fig. 1. HIV-1 Env and influenza HA sequence variability and glycosylation.
Sequence variability is represented on the molecular surface as varying colors described on the scale. Potential N-linked glycosylation sites from the consensus sequences are shown as purple spheres. The receptor binding site is colored in blue. (A) As no crystal structure of the full HIV-1 Env trimer is known, a model was generated from the electron microscopy reconstruction of the unliganded HIV-1 Env trimer (gray mesh, EMD ID 5019) (8), the gp120 core structure (PDB ID 3DNN) (8, 15), the gp120 mini-V3 loop (PDB ID 3TYG) (134), and the gp120 V1/V2 loops (PDB ID 3U4E) (17). Missing regions of gp120 (N- and C- termini, and the full V1/V2 and V3 loops) as well as the gp41 ectodomain are labeled inside brown and blue spherical shapes, respectively. (B) The influenza HA trimer structure was rendered using the coordinates from PDB ID 3GBN. This figure was prepared using Chimera (198).