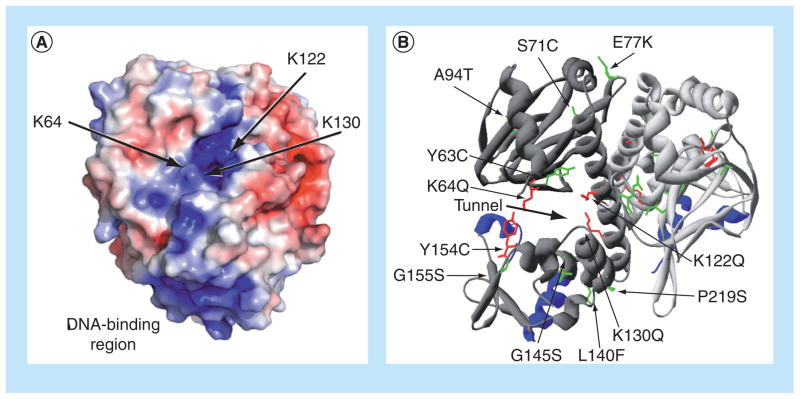
Figure 3. Location of the putative PrfA cofactor-binding pocket and of mutations that influence PrfA activation.
(A) Electrostatic modeling of wild-type PrfA protein demonstrating the potential distribution of solvent-accessible surface charges on the protein dimer and indicating binding-pocket mutations. Positive charge is shown in blue and negative charge is shown in red, with electrostatic potentials ranging from −4 kT/e (red) to +4 kT/e (blue). Arrows point to the lysine residues that contribute to the positive charge of the putative cofactor-binding pocket within PrfA. The positive charge of the DNA-binding region is also highlighted at the bottom of the PrfA monomer. (B) Ribbon modeling of PrfA, highlighting the putative cofactor-binding pocket described by Eiting et al. [62], as indicated by the thick black arrow, and identifying amino acid substitutions that influence PrfA activation. The monomers that make up the dimer are colored either light or dark gray, and the DNA-binding helix-turn-helix motifs are shown in blue. PrfA* mutations resulting in high levels of PrfA-dependent virulence gene expression are colored in green, while specific mutations abrogating or reducing PrfA activation are colored in red.
(A) Reproduced with permission from [74].