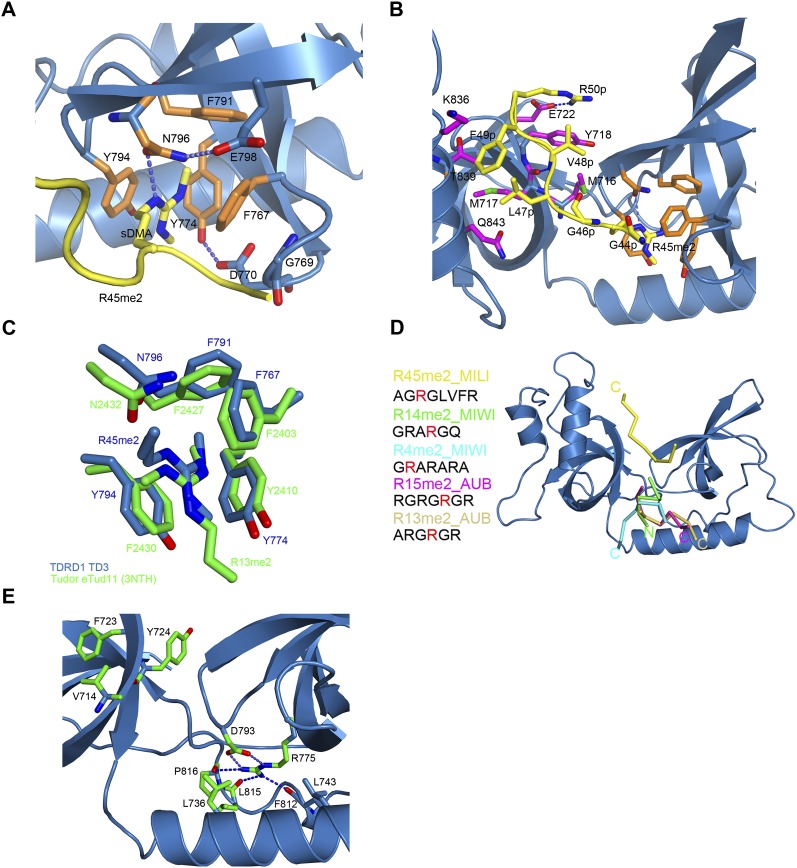
FIGURE 3.
TD3 binds the R45me2 peptide in a different orientation from that previously observed in eTud domains–peptide complexes. (A) The sDMA in the aromatic cage of TD3 colored as in Figure 2A. The dipeptide GD stabilizing the cage and E798 interacting with N796 are also shown. (B) Details of the interactions between the R45me2 peptide and TD3. Colors are as in A with additionally residues of the SN-like domain interacting with the peptide being in magenta. Blue dashed lines indicate putative hydrogen bonds. (C) Structural comparison of the aromatic cages of TD3 (blue) and eTud11 (green) in complex with their corresponding peptides highlighting the different direction of entry of the two methylarginine groups. All other such structures exhibit the same conformation as eTud11/R13me2. (D) Diagram showing the orientation of various sDMA-containing peptides with reference to the TDRD1 TD3 structure (blue) obtained by superposition of eTud–peptide complexes of known structure. R14me2 (PDB 3OMG, green) and R4me2 (PDB 3OMC, cyan) were co-crystallized with the SND1 protein, while R15me2 (PDB 3NTI, magenta) and R13me2 (PDB 3NTH, wheat) were crystallized with eTud11 from Drosophila TUDOR. All of the above-mentioned peptides enter the aromatic pocket from the “bottom,” while the R45me2 peptide (yellow) enters from the “top” of the structure. The observed peptide residues in each structure are shown at the left with the sDMA highlighted in red. N- and C-termini of the peptides are also indicated. (E) Conserved residues in murine TDRD eTud domains. In a stick-model representation, the residues conserved in most of the eTud domains of TDRD proteins are shown (green). The absolutely conserved R775 is involved in extensive interactions with residues of the Tudor domain, thus stabilizing the aromatic cage. Blue dashed lines indicate hydrogen bonds.