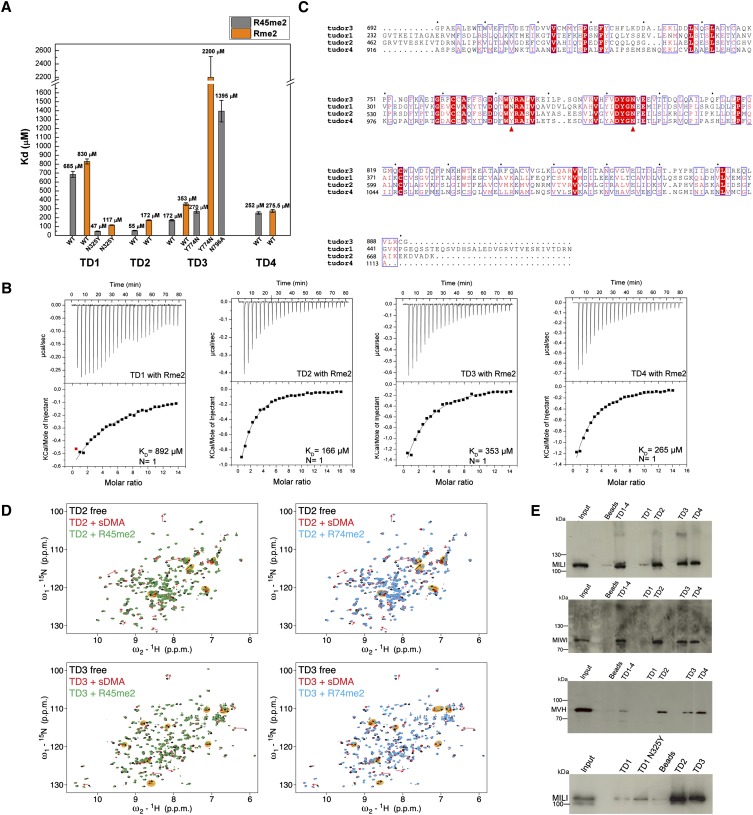
FIGURE 4.
“Active” and “inactive” extended Tudor domains of TDRD1. (A) KDs derived from ITC measurements for the binding of isolated symmetrically dimethylated arginine (Rme2) and R45me2 peptide to individual wild-type (wt) TDRD1 TDs and N325Y, Y774N, and N796A mutants. Error bars represent SD values from two experiments. (B) Representative ITC experiments and fits to the data. The four single eTud domains (TD1, TD2, TD3, and TD4) of TDRD1 with the isolated symmetrically dimethylated arginine (Rme2). (C) Multiple sequence alignment of the four individual Tudor domains of TDRD1. The first red arrow highlights the position of N325 of TD1, and the second arrow the Y774 of the aromatic cage of TD3. Mutations on both residues are critical for binding sDMAs (see A). (D) Binding of TD2 and TD3 domains of TDRD1 to sDMA-containing peptides monitored by NMR. Each panel shows an overlay of the 1H,15N HSQC spectra of the respective domain when free (black), when saturated with an excess of naked sDMA (red), and when saturated with MILI derived sDMA-containing peptides (green or cyan). For well-resolved peaks, chemical-shift perturbations arising from sDMA contacts are annotated with red arrows and those induced by interactions with flanking residues with black arrows in orange background. Notice that other peaks are affected by both sDMA and flanking residues but are not labeled. (E) Pull-down assays of endogenous murine proteins MILI, MIWI, and mouse Vasa homolog (MVH) by individual and multiple TDs of TDRD1. His-tagged constructs with four Tudor domains (TD1–4) and single Tudor domains (TD1, TD1 N325Y mutant TD2, TD3, TD4) were used. Size markers in kiladaltons are indicated.