FIGURE 5.
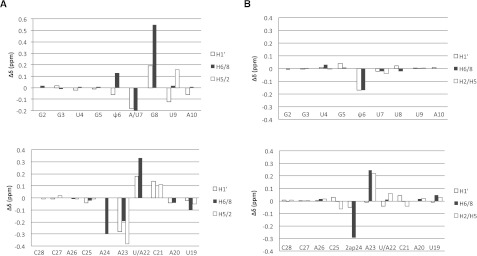
(A) Effects of the reverse UU2-Aintron base pair on the chemical shifts of the modified branch site duplex (mψBP and ψBP). Positive values for differences δ (mψBP) − δ (ψBP) are associated with downfield shifts of resonances in mψBP spectra as compared to those in ψBP spectra, whereas the negative values are associated with upfield shifts of respective resonances (U2 snRNA oligomer) (upper panel) and the intron oligomer (lower panel). Large changes in chemical shifts of mψBP relative to ψBP suggest perturbed helical parameters of the branch site region of mψBP. (B) Changes in chemical shifts of the modified ψ-dependent branch site duplexes as compared to the 2ap-substituted duplex (mψBP and mψBP2ap). Notation and orientation of respective strands are the same as in Figure 3. Changes in chemical shifts of nonexchangeable protons of ψ6, which may form a hydrogen bond with A23 or A24, are consistent with an intrahelical stacked conformation of 2ap24 in mψBP2ap.