Abstract
The human pathogenic yeast Candida glabrata harbors more than 20 surface-exposed, epithelial adhesins (Epas) for host cell adhesion. The Epa family recognizes host glycans and discriminates between target tissues by their adhesin (A) domains, but a detailed structural basis for ligand-binding specificity of Epa proteins has been lacking so far. In this study, we provide high-resolution crystal structures of the Epa1A domain in complex with different carbohydrate ligands that reveal how host cell mucin-type O-glycans are recognized and allow a structure-guided classification of the Epa family into specific subtypes. Further detailed structural and functional characterization of subtype-switched Epa1 variants shows that specificity is governed by two inner loops, CBL1 and CBL2, involved in calcium binding as well as by three outer loops, L1, L2, and L3. In summary, our study provides the structural basis for promiscuity and specificity of Epa adhesins, which might further contribute to developing anti-adhesive antimycotics and combating Candida colonization.
Keywords: molecular recognition, T-antigen, lectin, fungal pathogen
The strict mammalian commensal yeast Candida glabrata can act as a health care associated opportunistic pathogen. In immune-compromised patients, colonization results in lethal systemic infections and sepsis (2, 3). Besides that, C. glabrata is the causative agent of several less severe, mucosa-related, oral or urogenital diseases (4, 5). Due to its resistance against traditional antimycotics (6), this pathogen is considered to be highly emergent (4), and new therapeutic tools are being actively sought. One putative target for drug development is the epithelial adhesin (Epa) machinery, which is represented in C. glabrata by up to 23 different genes. Apart from EPA1, which confers adhesion of fungal cells to mammalian surfaces in vitro (7) and in vivo (8), the function of other EPA genes is only partly understood. For example, EPA6 and EPA7 have been identified as virulence factors, and EPA6 is further linked to biofilm formation and colonization of the vaginal epithelium (6). Interestingly, most of the EPA genes are clustered in subtelomeric regions, indicating rapid genetic adaptation to varying environmental conditions during host colonization (1). Also, expression of EPA1, EPA6, and EPA7 is regulated by a distinct silencing mechanism via the Sir machinery and mutation of SIR genes causes hypervirulence and enhanced kidney colonization (9).
Members of the Epa family are highly glycosylated mannoproteins with a modular architecture common to fungal GPI-anchored cell wall proteins (GPI-CWP) (10). While the N-terminal region (adhesion, or A domain) resembles C-type lectins mediating calcium-dependent adhesion to carbohydrate structures, the central serine- and threonine-rich segment (B domain) comprises a variable number of highly O-glycosylated repeats. Finally, a C-terminal anchor (C domain) cross-links the adhesin covalently to the cell wall (7, 11). Epa A domains are distantly related to flocculins from Saccharomyces cerevisiae, which also harbor A domains belonging to the PA14/Flo5-like family (12, 13). For the A domain of the flocculin Flo5, mannoside recognition involves a novel motif not found in other C-type lectins (12), the DcisD motif, whose role in galactose recognition by Epa1 has been recently confirmed (14). Other structural features determining sugar specificities by Flo5-like domains are so far unclear and do not allow correlating the different Epa proteins to their distinct and mostly still unknown specificities.
Recent, semiquantitative functional characterizations of Epa1, Epa6, and Epa7 showed that the specificity of these three adhesins is related (15). All three adhesins require a terminal galactose containing carbohydrate ligand for host recognition. Epa6 binds to oligosaccharides comprising galactose linked via 1–3 or 1–4 glycosidic bonds to glucose, galactose, or their N-acetylated derivates. Furthermore, Epa6 does not discriminate between α- and β-glycosidic linkages. In contrast, Epa1 has a narrower specificity, mainly excluding α-linked carbohydrates. Epa7 is the most specific of the three adhesins and almost exclusively binds to lactose (Galβ1–4Glc), Galβ1–3Gal, or their respective N-acetylated derivatives (15). Finally, adhesion specificity of EpaA domains is claimed to be governed by hypervariable stretches comprising amino acids 114–139 and 227–231 (15). However, a structural basis correlating these findings to Epa specificities has not been established.
Here, we present high-resolution crystal structures of the Epa1A domain complexed to cognate disaccharide ligands including Galβ1–3Glc or the T-antigen (Galβ1–3GalNAc). We further provide a structure-guided classification of the Epa family into specific subtypes and present structures of subtype-switched Epa1 variants with modified binding sites that correspond to Epa2, Epa3, and Epa6. Together with high-throughput carbohydrate-binding screens and fluorescence titration analyses these structures uncover the critical differences within the Epa protein family in regard to specificity and promiscuity. In vivo, subtype-specific binding of Epa proteins to epithelial surfaces in part depends on the specificity-determining loop region CBL2. Overall, Candida species can generate a large repertoire of Epa adhesins for colonizing various host niches by exerting subtle structural changes to a common motif.
Results and Discussion
Epa1A Preferentially Recognizes Galβ1–3 Glycans.
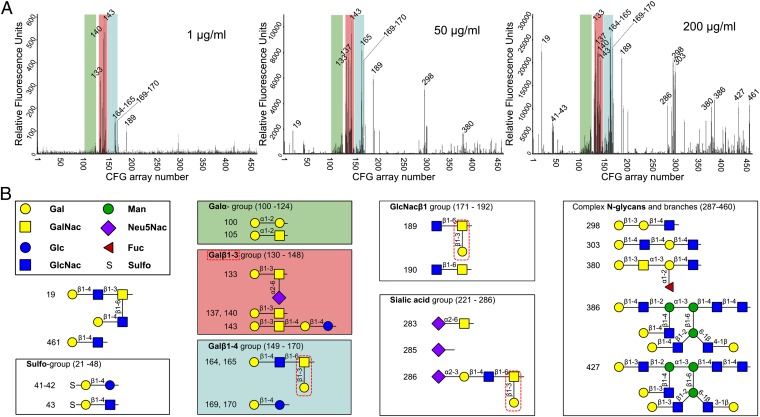
To determine the carbohydrate specificity profile of Epa1, the recombinant, fluorescence-labeled Epa1A domain was analyzed by glycan array screening [Consortium for Functional Glycomics (CFG)]. Binding of Epa1A to such arrays at 200 μg/mL showed a marked preference for glycans containing galactose at nonreducing ends (Fig. 1 A and B). Both α- and β-linked galactoses are bound by Epa1A, but the binding capability for Galβ1-terminated glycans is significantly higher than for those with Galα-terminal residues. Previous binding studies with Epa1A presented at the surface of S. cerevisiae indicated that this adhesin does not discriminate between mucin-relevant Galβ1–3 and Galβ1–4 disaccharides (15). Here, also no obvious preference was found when comparing Galβ1–3 and Galβ1–4-containing glycans, but all of the best binders within the Galβ1–4 group contain terminal Galβ1–3 branches (Fig. 1B). To further discriminate between stringent primary and nonstringent secondary specificities, binding at lower protein concentrations was conducted. At 50 μg/mL Epa1A, a moderate decrease in the group of Galβ1–4-containing glycans was observed that sharply drops at 1 μg/mL. Likewise, oligosaccharides with terminal α-linked galactose moieties show diminished binding at restricting Epa1A concentrations, further pointing to a preference for β-linkage. Best binders are Galβ1–3 group members, especially those with a Galβ1–3GalNAc motif including the T-antigen (glycans 137 and 140, Fig. 1A). In contrast, β1–4-linked lactosides, which are known to inhibit Candida adherence to host cells in the millimolar range (7), show only weak (lactose, glycans 169 and 170) or very moderate (N-acetyllactosamine, glycans 167 and 168) binding to Epa1A. In summary, our data provide a refined specificity profile for Epa1 and offer compelling evidence that this adhesin prefers Galβ1–3 disaccharides, but lacks high affinity for Galβ1–4 disaccharides. This conclusion is further supported by our fluorescence titration analysis (Fig. S1), which demonstrates that Epa1A has at least a 16-fold higher binding affinity for the T-antigen (KD = 2.1 ± 0.3 µM, Galβ1–3GalNAc) than for milk-derived lactose (34.6 ± 6.1 µM), which is likely to contain significant amounts of a Galβ1–3Glc contaminant as delineated by our structural analysis below.
Fig. 1.
Glycan binding by the Epa1A domain. (A) Binding profiles of Epa1A at different protein concentrations (1, 50, and 200 μg/mL), using the CFG array V4.1 harboring 451 different glycan structures. Relative fluorescence units as monitored reflect relative affinities toward the corresponding glycan. Glycans bound by Epa1A are indicated by their CFG array numbers. Overall, Epa1A recognizes exclusively terminal galactosides. Green, Galα group; red, Galβ1–3 group; blue, Galβ1–4 group. (B) Groups that are recognized by Epa1A are shown; their locations within the CFG array are given in parentheses. Structural formulas are described according to the CFG nomenclature. At 200 μg/mL, Epa1A recognizes both Galβ1–3 (red) and Galβ1–4 (blue) glycans. However, the latter can harbor terminal β1–3 ramifications (red dotted lines). At low concentrations, Epa1A has a much narrower specificity profile.
Overall Epa1A Structure.
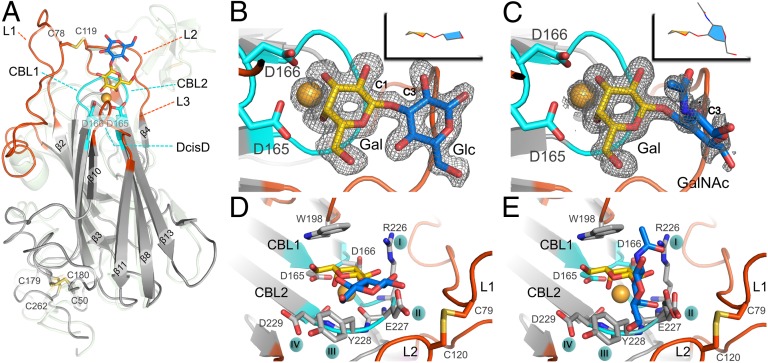
To determine the structure of Epa1A, crystals were obtained at low pH, but only in the presence of high concentrations of lactose. The Epa1A crystal structure was solved by molecular replacement, using a truncated homology model based on the S. cerevisiae Flo5A structure (12). Despite a pairwise sequence identity of less than 25%, the resulting structure (Tables S1 and S2) reveals a high similarity with Flo5A, as indicated by an rmsd of 1.4 Å over 121 common Cα atoms (Fig. 2A). The barrel-shaped Epa1A domain has an overall dimension of 57 × 42 × 36 Å3, consists of 15 β-strands, and is complexed via a calcium ion to its respective disaccharide ligand (Fig. 2A). Eleven of the strands form an antiparallel β-sandwich motif with PA14/Flo5-like topology (16). An L-shaped stretch, composed of the remaining four β-strands and the N and C termini, wraps around the β-sandwich and protects its bottom end from solvent access (12). This end of Epa1A is adjacent to the repetitive stalk of the Epa1 B domain. The two disulfide bridges C50–C179 and C180–C263, respectively, which covalently tether the N and C termini to the β-sandwich domain (Fig. 2A), are preserved in Flo5, suggesting that this rigid linkage between A and B domains is a general feature of GPI-CWP adhesins.
Fig. 2.
Glycan-binding site of the Epa1 adhesin. (A) Structure of the Epa1A domain (gray) with Galβ1–3Glc (Gal, yellow; Glc, blue). The outer subsite for glycan binding is formed by loops L1 and L2 (red), and the inner subsite comprises the CBL loops (cyan) and a Ca2+ ion (orange). The Flo5A structure is shown as a green transparent overlay, for comparison purposes. The Galβ1–3Glc (B) and T-antigen (C) ligands are shown within the Epa1A-binding site with their SIGMAA-weighted omit electron density maps, respectively (contouring level: 1σ, 0.37electrons/Å3 and 0.8σ, 0.31electrons/Å3). (D and E) Perpendicular views on the Epa1A-binding site either with the Galβ1–3Glc ligand (D) or with the T-antigen (E). Loops L1 and L2 are covalently linked via a cysteine bridge. W198 from L3 and Ca2+ interact directly with the galactose moiety, whereas the side chains from the CBL2 tip interact with glucose or GalNAc hydroxyls.
Which structural motifs define the ligand-binding site in Epa1A? In contrast to S. cerevisiae flocculins, Epa1A lacks the Flo5-like subdomain, which in budding yeast confers specific interactions with terminal α-1,2-mannobioside ligands (12). Surprisingly, the difference electron density map within the binding pocket of the Epa1A/lactose cocrystals clearly ruled out lactose, i.e., Galβ1–4Glc, as bound ligand. At 1.5 Å resolution only a Galβ1–3Glc disaccharide, a contaminant from commercially available lactose, was consistent with the electron density observed (Fig. 2B). This serendipitous finding corroborates the strong preference of Epa1A for Galβ1–3 over excess of Galβ1–4-linked disaccharides as indicated by our glycan profiling and fluorescence titration analyses. The binding site of the Galβ1–3 disaccharide is formed of five loops: two inner, short ones involved in calcium binding and ligand recognition (CBL1 and CBL2) and three outer, longer loops (L1, L2, and L3), which shield CBL1 and CBL2 from solvent access (Fig. 2D). CBL1 links β−strand 8 with 9 and is hallmarked by an unusual cis-peptide between D165 and D166, the DcisD motif. This motif is crucial for the C-type lectin function of Flo5A (12) and is conserved throughout the entire Epa family (Fig. S2). CBL2 links β-strands 12 and 13 and directly contributes to specific glycan binding by four of its side chains, R226, E227, Y228, and D229 (Fig. 2 D and E), which we further refer to as CBL2 positions I–IV (Fig. 3). Finally, the Ca2+-binding site comprises the side chain of N225, the carbonyl groups of peptide bonds within CBL2, and the carboxylic side chains of D165 and D166, the DcisD motif of CBL1. Moreover, the Ca2+ ion directly interacts with the nonreducing galactose moiety over the 3- and 4-hydroxyl groups (Fig. 2B) and is essential for glycan binding (Fig. S1).
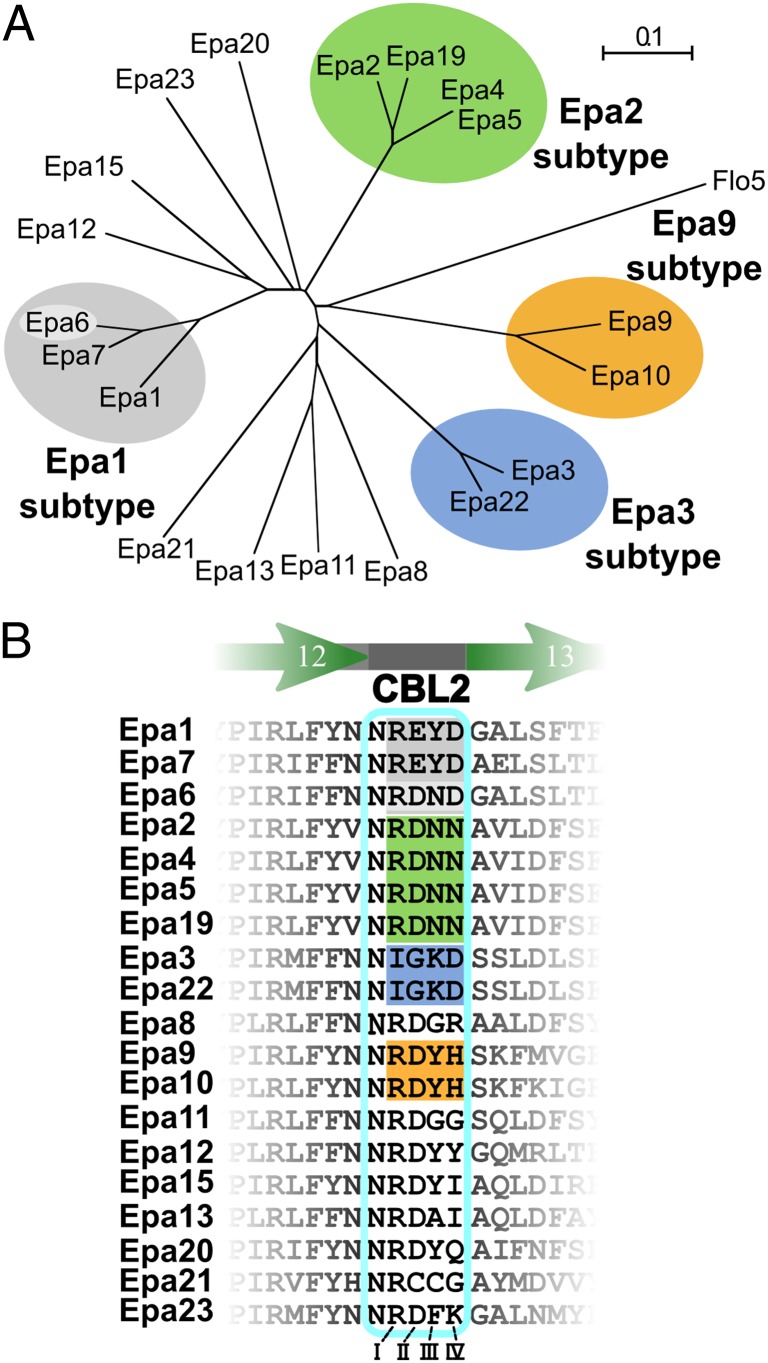
Fig. 3.
Phylogeny of C. glabrata EpaA domains. (A) Phylogenetic tree of EpaA domains. The tree is based on a structure-guided multiple-sequence alignment (Fig. S2), using the crystal structures of C. glabrata Epa1A and S. cerevisiae Flo5A (PDB ID: 2XJP). The Epa family shows four major, distinct subtypes: Epa1 (gray), Epa2 (green), Epa3 (blue), and Epa9 (orange). The scale bar indicates phylogenetic distances in number of amino acid substitutions per position. (B) Alignment of the CBL2 regions (cyan box). The Epa1, Epa2, Epa3, and Epa9 subtypes are color-shaded as in A. The Roman numerals indicate the four variable residues within CBL2.
Disaccharide Recognition by the Inner and Outer Subsites of Epa1A.
What is the structural basis for disaccharide discrimination by Epa1A? The Epa1A⋅Galβ1–3Glc complex indicates that at least two subsites are crucial for glycan recognition: (i) the inner subsite, which is formed of CBL1, the Ca2+ ion, R226 from CBL2, and W198 from loop L3 and confers specific binding to the terminal galactose, and (ii) the outer subsite, which consists of the side chains of CBL2 together with loops L1, L2, and L3 and interacts with the peripheral hexose moiety (Fig. 2 A and C). Moreover, loop L1 is not part of the core PA14/Flo5A-like domain, but connects β-strands 2 and 3. Also, a disulfide bridge formed by C78 and C119 cross-links L1 and L2 to arrest their conformation for shielding the binding site from bulk solvent.
The Epa1A⋅Galβ1–3Glc complex further reveals that the galactose moiety in the inner subsite adopts a flipped orientation, which differs from the orientation of mannose in Flo5A (12). This flipped binding mode is crucial for coaligning the pyranose moiety with residue W198 from loop L3 as well as for hydrogen bonding between the R226 side chain and the 2- and 3-hydroxyls of galactose (Fig. 2D). This suggests that the indole side chain of W198 together with R226 is crucial for selecting the terminal galactose (Figs. S3 and S4), a conclusion that is supported by our finding that an Epa1W198A variant shows only marginal binding activity (Fig. S5). We also performed molecular dynamics studies for ligand binding by Epa1A (Movies S1, S2, and S3), which indicate that W198 may exert an organizing effect on loop L3, because they show that the stretch W198–T202 of loop L3 adopts the observed degree of order only upon complexation of galactose.
The outer subsite apparently selects the type of glycosidic bondage and nature of the hexose moiety linked to the terminal galactose. The Epa1A⋅Galβ1–3Glc structure shows that the site for recognition of Galβ1–3-linked hexoses not only comprises E227 and Y228 of CBL2 (Fig. 2 B and D), but also loops L1 and L2 lining the outer site. The glutamic acid side chain projected from position II forms hydrogen bonds with the 2-hydroxyls of both hexoses of Galβ1–3Glc as well as with loop L2 via the peptide group of G118–C119. Likewise, the L2 loop wraps with A115–G118 around Y228 and thereby stabilizes its packing with the pyranose ring of the glucose within the Galβ1–3Glc ligand.
To obtain further insights into disaccharide binding specificity, the structure of the cognate Epa1A⋅T-antigen complex was solved at 1.24 Å resolution. This structure reveals the same mode of glycan recognition at its inner subsite as found for the Epa1A⋅Galβ1–3Glc complex, but major differences at the outer subsite (Fig. 2 C and E). Whereas in the Epa1A⋅Galβ1–3Glc complex the glucose moiety is coplanar to galactose, the N-acetylgalactosamine moiety of the T-antigen adopts an orthogonal conformation when complexed to Epa1A. This diverging conformation is apparently caused by the hydrogen bond between the axial 4-hydroxyl group and E227 of CBL2. As shown above, the Epa1A binding site is highly specific by disfavoring Galβ1–4-linked glycans. Molecular modeling of a lactose complex and subsequent molecular dynamics studies show that a β1–4 linkage fails to position the second hexose in a defined conformation within the outer subsite, e.g., by packing with Y228 (Fig. S4 and Movies S3 and S4). Nevertheless, the outer subsite harbors sufficient flexibility to allow different binding modi, as shown for the T-antigen. This suggests that EpaA domains permit a certain degree of promiscuity for disaccharide binding in their outer, but not in their inner ligand-binding subsite.
Differential Promiscuity of Epa Subtypes Is Dominated by CBL2.
We next generated a sequence alignment of 19 EpaA domains that is based on the structures of Epa1A and Flo5A (Fig. S2) and provides a structure-guided phylogenetic tree of the Epa family (Fig. 3A). Four prominent branches appear in this tree, which cluster 11 EpaA domains into four subtypes, which were named Epa1 (Epa1A, Epa6A, Epa7A), Epa2 (Epa2A, Epa4A, Epa5A, Epa19A), Epa3 (Epa3A, Epa22A), and Epa9 (Epa9A, Epa10A), respectively. Remarkably, the CBL2 positions I–IV are mostly sufficient to discriminate between the different subtypes (Fig. 3B). For Epa1 subtype members, this motif consists of REYD or RDND, and the Epa2 and Epa9 subtype adhesins contain related RDNN or RDYH motifs. Only Epa3 subtype proteins show a highly diverging IGKD motif. In addition, the Epa9 subtype differs from others by a prominent elongation within the L1 loop.
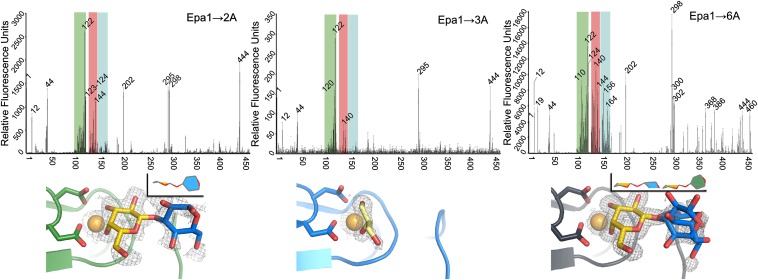
To explore how CBL2 affects the specificity of different Epa subtype proteins, the CBL2 motif of Epa1A was exchanged for the motif of Epa2A and Epa3A (Epa1→2A and Epa1→3A variants). Because the Epa1 subtype includes two different CBL2 signatures, an Epa1→6A variant was also generated. However, the Epa9 subtype was not further investigated due to its diverging L1 loop. In the next step, fluorescence titration analysis of the subtype-switched Epa1A variants was performed. This analysis revealed that the binding affinities of the Epa1→6A variant for lactose and the T-antigen resemble those of Epa1A (KD = 33.5 ± 6.6 µM and 1.7 ± 0.4 µM). In contrast, the Epa1→2A variant has a fourfold lower affinity for lactose (KD = 128 ± 16.3 µM). Unfortunately, this variant rapidly aggregates in the presence of the T-antigen, thus preventing quantification of binding despite its obvious interaction with this glycan. For the Epa1→3A variant, we detected only extremely low affinity toward lactose (KD = 27.3 ± 5.7 mM) and a 15-fold less efficient binding of the T-antigen (KD = 30.0 ± 6.1 µM) when compared to Epa1. To further explore the role of the CBL2 motif, the subtype-switched Epa1A variants were subjected to CFG glycan arrays, revealing further differences in their ligand-binding specificity (Fig. S6). In contrast to Epa1, the Epa1→6A variant was found to recognize glycans within the Galβ1–3 group without clear discrimination between α- and β-galactosides (Fig. 4). In addition, Epa1→6A weakly prefers glycans harboring β1–3-linked galactose moieties within the Galβ1–4 group, with glycans 164 and 165 found as best binders, as well as Galβ1–4 terminated glycans like N-acetyllactosamine (glycan 168). As found in the fluorescence titration analysis, lactose is bound with much weaker affinity by this variant, similar to Epa1 (Fig. 4). For Epa1→2A and Epa1→3A, we found strongly diminished binding compared to Epa1A and Epa1→6A, which were highly biased toward glycans with terminal, α-linked galactose moieties. In summary, all variants analyzed by glycan array profiling are highly specific for galactose in the inner subsite, whereas the outer subsite is promiscuous, being occupied by glucose, N-acetylglucosamine, galactose, or N-acetylgalactosamine (Figs. S6 and S7), a finding that explains previous observations (15).
Fig. 4.
Specificity profiles and binding pockets of subtype-switched Epa1A variants. (Upper) CFG glycan array profiles for Epa1A variants at 200 μg/mL protein concentration. Strong binders are indicated by their CFG array number. Unlike Epa1A, Epa1→6A lacks the strict Galβ1–3 specificity and instead binds galactose-comprising glycans within the Galα, Galβ1–4, and Galβ1–3 groups (green, blue, and red regions, respectively), indicating increased promiscuity. Epa1→2A and Epa1→3A have narrowed specificity profiles with Epa1→3A preferring terminal α-joined galactose-comprising glycans. The lower fluorescence signals of Epa1→2A and Epa1→3A indicate weaker binding to glycans than that of Epa1A and Epa1→6A. (Lower) Top view of subtype-switched Epa1A-binding sites with SIGMAA-weighted omit electron densities for ligands complexed to Epa1→6 (anthracite gray, contouring level: 0.8σ, 0.29electrons/Å3), Epa1→3A (blue, 0.8σ, 0.28electrons/Å3) and Epa1→2A (green, 0.6σ, 0.19electrons/Å3). The orientation of the disaccharide within the binding pocket is shown (Upper Right Inset). CBL loops of the inner subsite are marked in cyan and L loops of the outer subsite in orange.
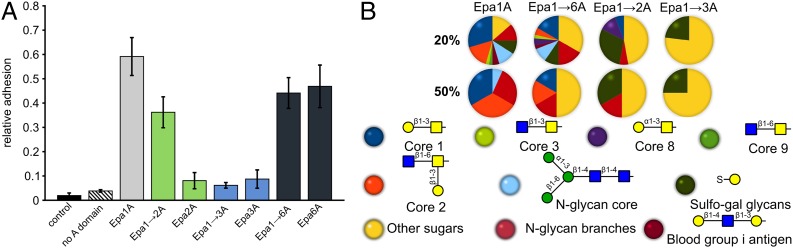
Next, the adhesion behavior of different Epa subtypes was analyzed in vivo, using a heterologous S. cerevisiae system presenting EpaA domains on the cell surface (Fig. S8) and the human colorectal, epithelial cell line Caco-2. The T-antigen is a typical mammalian surface glycan, which is present at high densities on Caco-2 cell surfaces, but not on fungal cells. As expected, S. cerevisiae strains that lack an EpaA domain failed to adhere to epithelial cells (Fig. 5A). In contrast, strains presenting Epa1A or subtype-switched Epa1→6A strongly adhere to Caco-2 cell layers. Obviously, T-antigen–presenting epithelial cells are well recognized by the Epa1 subtype independently of the used sequence of its CBL2 motif (Fig. 5A). Furthermore, the 15-fold lower affinity of Epa1→3A to recognize the T-antigen in vitro is mirrored in vivo, because strains presenting either Epa1→3A or native Epa3 only inefficiently bind to Caco-2 cells (Fig. 5A). Epa1→2A that clearly mediates binding to Caco-2 cells presents a different scenario. Given an only fourfold reduced in vitro binding affinity to lactose compared to Epa1A and Epa1→6A (Fig. S1), one may expect that the affinity or at least koff of Epa1→2A against the T-antigen is similar to that of the Epa1A subtypes. However, the native Epa2A domain fails to confer significant epithelial cell binding, suggesting that in this case switching the specificity and binding characteristics between Epa subtypes seems to require more than exchanging the CBL2 motif. Additional structural features of the outer subsite like the L1 and L2 loops may hence contribute to binding and specificity. Nevertheless, our results indicate that the CBL2 loop plays a prominent role in conferring specificity and promiscuity to Epa family adhesins and thereby significantly affects efficient host cell recognition.
Fig. 5.
In vivo binding of EpaA domains and the distribution of mucin-derived glycan ligands. (A) Relative adhesion to epithelial cells as conferred by different EpaA domains was determined by using a heterologous S. cerevisiae expression system. S. cerevisiae strains presenting different EpaA domains on Flo11 BC stalks were incubated with a monolayer of Caco-2 cells and adhesion was determined after 2 h. Colors correspond to the different EpaA subtypes as shown in Fig. 3A. (B) Pie charts show glycan types of the CFG array for which binding signals exceed either 20% or 50% of the signal of the best binder. For each glycan type (color as above), the core structures are shown below in CFG standard notation. Nonassigned glycans are grouped as “other sugars” (yellow).
Subtle Structural Changes Cause Promiscuity.
To obtain further detailed insights into Epa protein functionality, the structures of the subtype-switched Epa1A variants were determined by using the orthorhombic crystal form obtained from cocrystallization with lactose. This analysis revealed structural alterations of the Epa1A variants, which were restricted to their glycan-binding sites when compared to Epa1 (Fig. 4). Similar to Epa1, the Epa1→2A and Epa1→6A variants were found to harbor the Galβ1–3-linked disaccharide with a well-defined positioning of the galactose moiety in the inner subsite. In contrast to Epa1, however, the terminal glucose residue was present in two alternative conformations within the outer subsite of Epa1→6A (Fig. 4). This suggests that the changes at positions II and III in CBL2 (E227D, Y228N) cause a wider and more flexible outer subsite when compared to that in Epa1A (Fig. 4). As a consequence, the reducing end of the disaccharide can almost freely rotate due to increased entropic contribution to binding. Thus, sterically demanding glycans appear to be more easily accommodated in Epa6-like binding sites, a conclusion that is supported by our glycan profiling, which shows increased promiscuity for Epa1→6A, because it efficiently binds to Galβ1–3 and Galβ1–4 as well as to Galα joined glycans (Fig. 5B and Fig. S6C). Interestingly, Epa6 not only confers epithelial cell adhesion, but also is involved in the formation of biofilms, a prerequisite for C. glabrata to colonize inert surfaces such as clinical catheters (17, 18). The reduced Epa6 specificity and tendency to biofilm formation may thus promote host persistence due to improved resistance to antifungal agents. Accordingly, Epa6 has been assigned a role in urinary tract infections (19).
In the Epa1→2A⋅Galβ1–3Glc complex, the glucose adopts an orthogonal conformation similar to that of the Epa1→6A⋅Galβ1–3Glc complex (Fig. 4). Having the same exchanges at CBL2 positions II and III, the Epa1→2A variant recognizes Galα-joined glycans as well. In addition, the change in position IV (D229N) reduces the negative charge next to the galactose ligand. As a consequence, Epa1→2A exhibits a marked preference for terminal galactosides that are sulfated at the 6-hydroxyl position (Fig. 5B). These glycan structures are prevalent in sulfomucins, which are in turn predominant components of intestinal mucosa (20). The structure of the Epa1→2A⋅Galβ1–3Glc complex offers sufficient room in the inner subsite to position the 6-sulfate of these terminal sulfo-galactosides close to N229. In contrast, the Epa1 subtype is hindered to recognize sulfo-galactosides due to possible electrostatic repulsion by D229 at position IV.
Finally, the Epa1→3A variant that is strongly impaired for in vitro and in vivo ligand binding harbors a glycerol molecule in its binding pocket that stems from the cryo buffer (Fig. 4). The glycerol is coordinated to Ca2+ via two vicinal hydroxyl groups similar to the galactose moiety in Epa1. The Epa3 subtype is characterized by a highly divergent CBL2 region with positions I–III being replaced by IGK. Apparently, a change at position I from arginine to isoleucine (R226I) is sufficient to weaken the interaction with galactose in the inner subsite and to cause impaired discrimination of glycerol from hexoses (Fig. S6B). Nevertheless, Epa1→3A still has residual affinity toward the T-antigen, i.e., 15-fold lower than that of other Epa1A variants (Fig. 4). Finally, Epa1→3A binds more efficiently to 6-sulfated galactosides than Epa1. This indicates that the lysine residue at position III of the Epa3 subtype CBL2 is suitably positioned to form a salt bridge with the 6-sulfate of the sulfo-galactoside bound to the inner subsite. This may overcome the electrostatic interference by aspartate at position IV that is observed for the Epa1 subtype.
Conclusions
Our detailed structural and functional characterization of Epa1A and subtype-switched variants demonstrates that the specificity of C. glabrata Epa adhesins for different glycan structures is exerted by a distinct region of their A domain. This region comprises the calcium-coordinating loops CBL1 and CBL2 as well as parts of loop L3 and bears modular characteristics to generate Epa subtype variability. The specificity profiles of Epa1A and its variants strengthen and refine the link between these C. glabrata epithelial adhesins and the cores of mucin-type O-glycans (15). Importantly, we found that the T-antigen, which represents the core 1 of mucin-type O-glycans (21) and is part of most mucin-derived glycans (22), is one of the best ligands for all EpaA variants investigated in this study. Other glycans that we found to be recognized by Epa1A and variants are also related to mucins and include other mucin-type O-glycan cores or N-glycans, e.g., sulfo-galactose–containing glycans.
Our study further reveals that the structural basis for the specificity and promiscuity in Epa–glycan interactions is significantly affected by the properties of CBL2. This finding is consistent with previous domain-swapping experiments between Epa6 and Epa1/7, which identified a larger, CBL2-covering region for controlling specificity (15). Specificity of the inner subsite for galactose is dictated by recognition of the cis-diol formed by the 3- and 4-hydroxyls. Furthermore, position I of CBL2 is occupied by an arginine in all Epa subtypes except Epa3. This crucial residue forms two hydrogen bonds with the 2-hydroxyl of the bound galactose and packing to loop L1 via W79 and Y83 stabilizes its conformation. In the outer subsite, position III of CBL2 presents either large aromatic residues (Y, Epa1, Epa9, Epa10, Epa12, Epa15, and Epa20; F, Epa23) or polar amino acids like asparagine (Epa2, Epa4, Epa5, Epa6, and Epa 19). Whereas the former promote packing with planar glucose/galactose derivatives, resulting in specific interactions with either terminal Galβ1–3Glc(NAc) or Galβ1–3Gal(NAc), the latter can increase promiscuity (Epa6) or enable additional binding to, e.g., sulfated galactoses (Lys: Epa3, Epa22). Position IV appears to mainly control the degree of allowed modification in the terminal galactose moiety. Together, CBL2 positions I–IV may be proposed to form a simple structural code for Epa specificity. However, the differences found for binding to host cell monolayers by Epa1→2A and Epa2A as well as the high-resolution structure of the Epa1A⋅T-antigen complex with its orthogonal binding mode for the GalNAc moiety challenge this notion and imply that the periphery formed by the loops L1 and L2 adds to the specificity of Epa adhesins.
In summary, our study suggests that variable presentation of Epa family members with diversified outer subsites at the cell surface of C. glabrata is one of the key elements for efficient and tissue-specific host invasion and could explain the clinical behavior of this human pathogenic fungus (23). Our structure-based insights that Candida–host interaction crucially depends on the conserved nature of an inner galactose-binding site of Epa adhesins may further contribute to the development of tailored antimycotics to combat this emerging pathogen.
Methods
Protein Production and Purification.
Recombinant Epa1A and variants were generated in a thioredoxin and glutathione reductase-deficient Escherichia coli strain. Epa1A was purified by NiNTA-affinity and size-exclusion chromatography.
Phylogenetic Analysis.
Phylogenetic analysis was performed by the neighbor-joining method, using Clustal X2.0 (24), COBALT (25), or a local copy of t-coffee (26) implemented with 3DCoffee (27) for structure-based alignments. Preliminary targets were selected with the help of BLAST.
Crystallization and Structure Determination.
Epa1A/lactose cocrystals belonging to space group C2221 were obtained from ammonium sulfate-containing conditions. Epa1A⋅T-antigen cocrystals were obtained by soaking Ca2+-stripped Epa1A/lactose cocrystals. The structure of Epa1A was then solved by molecular replacement and refined using REFMAC5 or PHENIX.
High-Throughput Glycan-Binding Assays.
The CFG glycan array consists of different groups of oligosaccharides that are presented by mammalian cells. Recombinant, fluorescently labeled Epa1A domains were applied to the CFG array V4.1 chips at concentrations ranging from 1 μg/mL to 200 μg/mL. Chip surfaces were repeatedly washed; remaining fluorescence was measured and quantified.
Fluorescence Titrations.
Fluorescence titrations of Epa1A and variants were performed against lactose and the T-antigen. Binding was followed at an emission wavelength of 346 nm by excitation of intrinsic tryptophan fluorescence at 295 nm. Fluorescence quench was recorded during titration and fitted using a one-site plus unspecific-binding model.
In Vivo Adhesion Assays.
In vivo adhesion of Epa proteins to epithelial cells was determined using a nonadhesive S. cerevisiae strain presenting the different EpaA domains at the cell surface (Tables S3–S5). Adhesion assays were performed as previously described (28).
Supplementary Material
Acknowledgments
The authors thank Tobias Klar and Alexander Popov for support at the European Synchrotron Radiation Facility, Grenoble (beamlines ID14-4 and ID23); Petra Gnau and Lisa Ludewig for technical support; Uwe Linne for mass-spectrometric analyses; and David Smith and the Consortium for Functional Glycomics for performing in vitro screening of glycan specificity (Consortium for Functional Glycomics Request 2080). This research is supported by Grants ES152/7, MO 825/1-4, and GRK 1216 from the Deutsche Forschungsgemeinschaft and by the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) Center for Synthetic Microbiology (H.-U.M. and L.-O.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AF9, 4AFA, 4AFB, 4AFC, and 4ASL).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207653109/-/DCSupplemental.
References
- 1.Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Gulia J, Aryal S, Saadlla H, Shorr AF. Healthcare-associated candidemia--a distinct entity? J Hosp Med. 2010;5:298–301. doi: 10.1002/jhm.652. [DOI] [PubMed] [Google Scholar]
- 3.Komshian SV, Uwaydah AK, Sobel JD, Crane LR. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: Frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;11:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 4.Buitrón García-Figueroa R, Araiza-Santibáñez J, Basurto-Kuba E, Bonifaz-Trujillo A. Candida glabrata: An emergent opportunist in vulvovaginitis. Cir Cir. 2009;77:423–427. [PubMed] [Google Scholar]
- 5.Pignato S, et al. Persistent oral and urinary Candida spp. carriage in Italian HIV-seropositive asymptomatic subjects. J Prev Med Hyg. 2009;50:232–235. [PubMed] [Google Scholar]
- 6.Mundy RD, Cormack B. Expression of Candida glabrata adhesins after exposure to chemical preservatives. J Infect Dis. 2009;199:1891–1898. doi: 10.1086/599120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- 8.Filler SG. Candida-host cell receptor-ligand interactions. Curr Opin Microbiol. 2006;9:333–339. doi: 10.1016/j.mib.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Castaño I, et al. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- 10.Linder T, Gustafsson CM. Molecular phylogenetics of ascomycotal adhesins--a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet Biol. 2008;45:485–497. doi: 10.1016/j.fgb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- 12.Veelders M, et al. Structural basis of flocculin-mediated social behavior in yeast. Proc Natl Acad Sci USA. 2010;107:22511–22516. doi: 10.1073/pnas.1013210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigden DJ, Mello LV, Galperin MY. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem Sci. 2004;29:335–339. doi: 10.1016/j.tibs.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Ielasi FS, Decanniere K, Willaert RG. The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: Structural and functional study of the carbohydrate-binding domain. Acta Crystallogr D Biol Crystallogr. 2012;68:210–217. doi: 10.1107/S0907444911054898. [DOI] [PubMed] [Google Scholar]
- 15.Zupancic ML, et al. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol Microbiol. 2008;68:547–559. doi: 10.1111/j.1365-2958.2008.06184.x. [DOI] [PubMed] [Google Scholar]
- 16.Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 17.Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- 18.Iraqui I, et al. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol Microbiol. 2005;55:1259–1271. doi: 10.1111/j.1365-2958.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- 19.Domergue R, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- 20.Nieuw Amerongen AV, Bolscher JG, Bloemena E, Veerman EC. Sulfomucins in the human body. Biol Chem. 1998;379:1–18. doi: 10.1515/bchm.1998.379.1.1. [DOI] [PubMed] [Google Scholar]
- 21.McEntyre J, et al. 2009. Essentials of Glycobiology. Cold Spring Harbor Lab Press, Cold Spring Harbor, NY), 2nd Ed.
- 22.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arendrup MC. Epidemiology of invasive candidiasis. Curr Opin Crit Care. 2010;16:445–452. doi: 10.1097/MCC.0b013e32833e84d2. [DOI] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulos JS, Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 26.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 27.O’Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C. 3DCoffee: Combining protein sequences and structures within multiple sequence alignments. J Mol Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 28.Dieterich C, et al. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology. 2002;148:497–506. doi: 10.1099/00221287-148-2-497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.