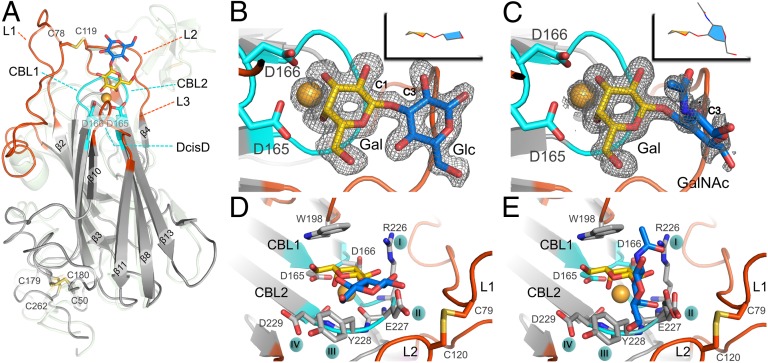
Fig. 2.
Glycan-binding site of the Epa1 adhesin. (A) Structure of the Epa1A domain (gray) with Galβ1–3Glc (Gal, yellow; Glc, blue). The outer subsite for glycan binding is formed by loops L1 and L2 (red), and the inner subsite comprises the CBL loops (cyan) and a Ca2+ ion (orange). The Flo5A structure is shown as a green transparent overlay, for comparison purposes. The Galβ1–3Glc (B) and T-antigen (C) ligands are shown within the Epa1A-binding site with their SIGMAA-weighted omit electron density maps, respectively (contouring level: 1σ, 0.37electrons/Å3 and 0.8σ, 0.31electrons/Å3). (D and E) Perpendicular views on the Epa1A-binding site either with the Galβ1–3Glc ligand (D) or with the T-antigen (E). Loops L1 and L2 are covalently linked via a cysteine bridge. W198 from L3 and Ca2+ interact directly with the galactose moiety, whereas the side chains from the CBL2 tip interact with glucose or GalNAc hydroxyls.