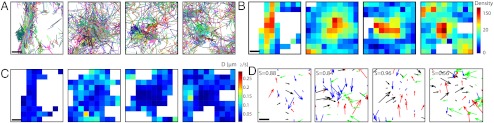
Fig. 5.
Analysis of superresolution trajectories of vesicular stomatitis virus G (VSVG) proteins. (A) Four samples of sptPALM trajectories (n = 30,000) of VSVG proteins. (B) Density map of the VSVG proteins containing high density areas. (C) Diffusion coefficient maps (computed from [3]). Low diffusion regions are colocalized with high protein density (red squares). (D) Field of forces in the four squares. No potential wells can be detected (the average index S is very high Savg = 0.81), showing that proteins do not interact at potential wells. (Scale bars, 200 nm).