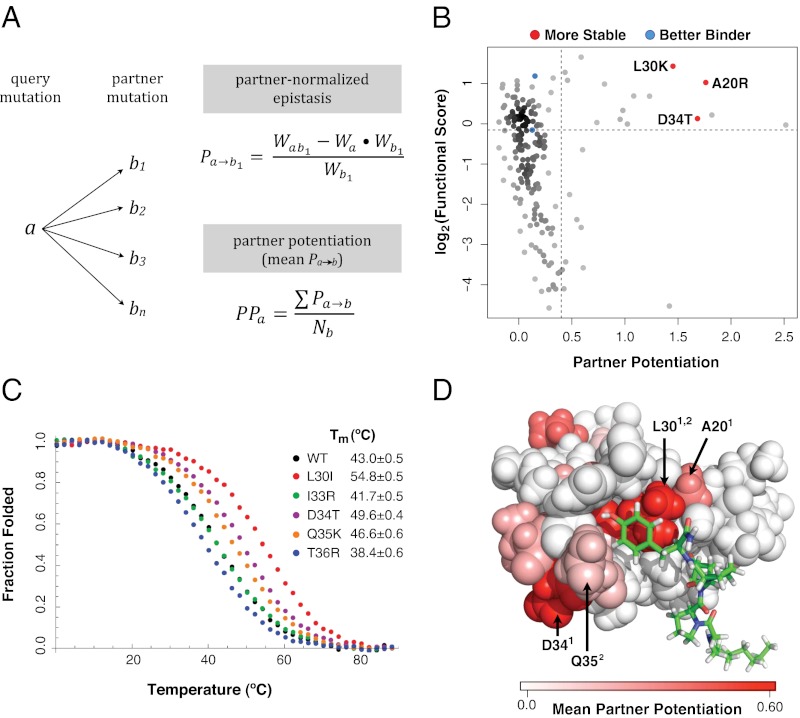
Fig. 3.
Partner potentiation reveals stabilizing mutations. (A) Partner potentiation is calculated for a query mutation a that forms doubly mutated variants with mutations b1, b2, b3 … bn. Partner-normalized epistasis scores are calculated for a by dividing the epistasis score of each double-mutant combination that a is found in by the functional score of the partner mutation (an example calculation for b1 is shown). The partner potentiation of a is calculated as the mean of the normalized epistasis scores (in this case, the scores for b1 … bn). (B) Partner potentiation is plotted for each single mutation against its functional score. The known stabilizing (A20R, L30K, and D34T) and activity-enhancing (K21R and Q35R) mutations are highlighted in red and blue, respectively. Mutations with a partner potentiation score greater than 0.4 and a functional score greater than 0.9 were considered to be stabilizing. (C) Stabilizing mutations were validated by thermal denaturation. The WT hYAP65 WW domain, the known stabilizing mutant D34T, and four candidate stabilizing mutants (L30I, I33R, Q35K, and T36R) are shown in black, purple, red, green, orange, and blue, respectively. (D) Positions in the hYAP65 NMR structure (Protein Data Bank ID code 1k9q) with stabilizing variants as judged by partner potentiation are shown, colored by the magnitude of the mean partner potentiation score using the PyMol software. Stabilizing mutations are distributed throughout the WW domain. Positions of previously known (superscript 1) and previously unknown (superscript 2) validated stabilizing mutations (20, 30, 34, 35) are highlighted.