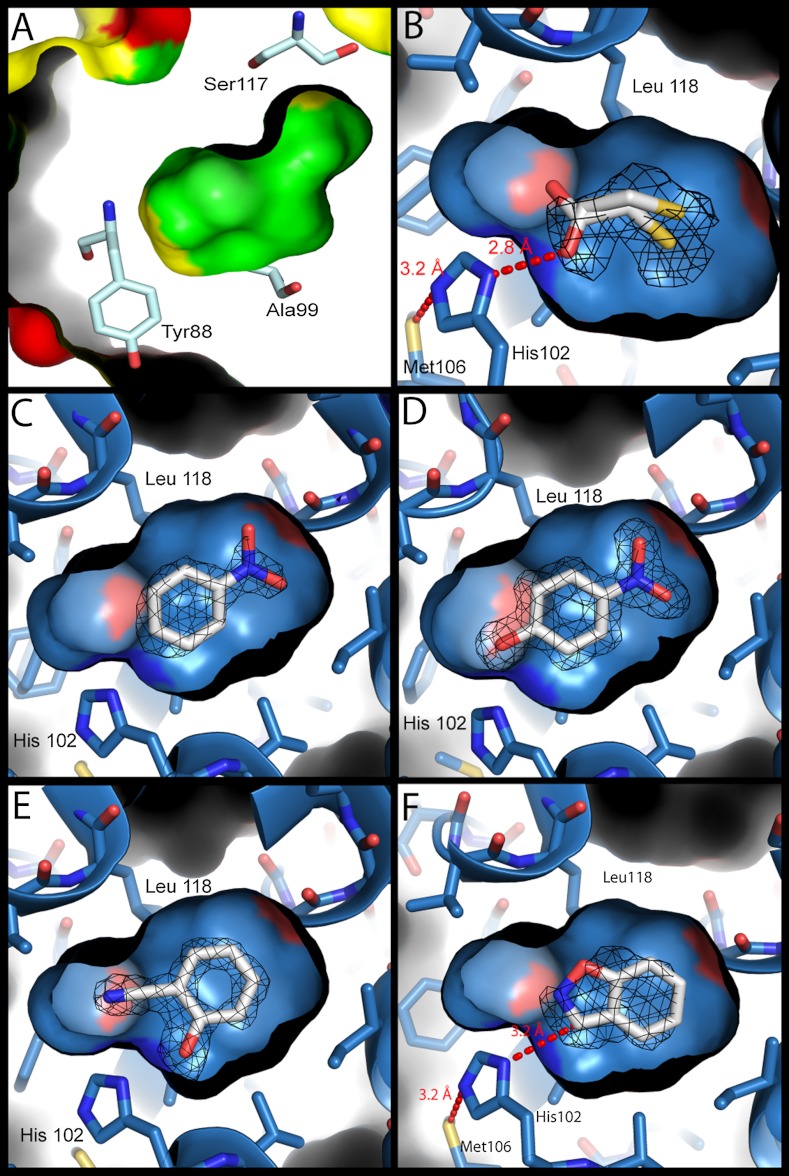
Fig. 2.
Structures of the model cavity sites. For panels B–F, the structures of the L99A/M102H† cavity site are shown as a blue surface with bound ligands (carbons white, nitrogens blue, oxygens red, sulfur yellow). Fo - Fc density (calculated after refinement but before the introduction of the ligand) at 3σ is shown as black mesh. (A) The molecular surface of the L99A cavity (PDB ID 181L) (inner) embedded within the overall surface of the protein (outer). The surfaces of ionic residues are shown in red, polar residues in yellow and nonpolar residues in green. Several cavity lining residues are rendered as sticks (oxygens red, nitrogens dark blue, carbons pale blue). The polar atoms of Tyr88 and Ser117 are oriented away from the cavity, which is almost entirely apolar in nature. (B) L99A/M102H† complexed with 2-mercaptoethanol at 1.30 Å. The hydrogen bond observed in all the structures between Sδ of Met106 and Nε2 of His102 is shown. (C) L99A/M102H† complexed with nitrobenzene at 1.64 Å. (D) L99A/M102H† complexed with 4-nitrophenol at 1.54 Å. (E) L99A/M102H† complexed with 2-cyanophenol at 1.49 Å. (F) L99A/M102H† complexed with benzisoxazole at 1.64 Å, modeled in the catalytically competent pose showing the 3.3 Å distance between Nδ1 of His102 and the acidic carbon of the Kemp substrate. Figures rendered with PyMol.