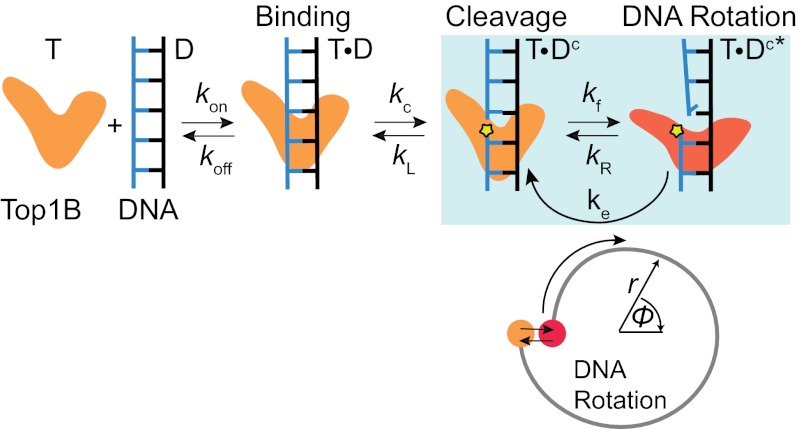
Fig. 6.
Proposed kinetic clutch model for Top1B. In this scheme, Top1B (T) reversibly binds DNA (D), forming a binary complex (T⋅D) in which the DNA is cleaved at the rate kc. The cleaved complex is initially in a religation-competent state (T⋅Dc) separated from the rotation-competent state (T⋅Dc*) by a torque-independent energy barrier, corresponding to a protein conformational change or a radial DNA distortion (orange and red states). The Top1B–DNA complex fluctuates between the religation-competent (T⋅Dc) and rotation-competent (T⋅Dc*) states at the torque-independent rates kR and kf. From the religation-competent state T⋅Dc, the DNA can be religated at rate kL. DNA rotation corresponds to escape from the T⋅Dc* state at the torque-dependent rate ke. Each rotation of the DNA corresponds to one cycle of T⋅Dc to T⋅Dc* and back to T⋅Dc as denoted by the blue-shaded box. The corresponding reaction coordinates are schematically represented below. The free end of the DNA rotates from the state T⋅Dc* (red dot) but is prevented from further rotation once it enters the state T⋅Dc (orange dot). A reversible radial motion (i.e., crossing an energy barrier orthogonal to rotation) is required to return the complex to the rotation-competent state. Although the model is based on positive supercoil relaxation, we speculate that a similar mechanism holds for negative supercoil relaxation. Based on the “intercalated model,” CPT likely binds Top1B in the religation-competent state (T⋅Dc) in which the two DNA ends are aligned.