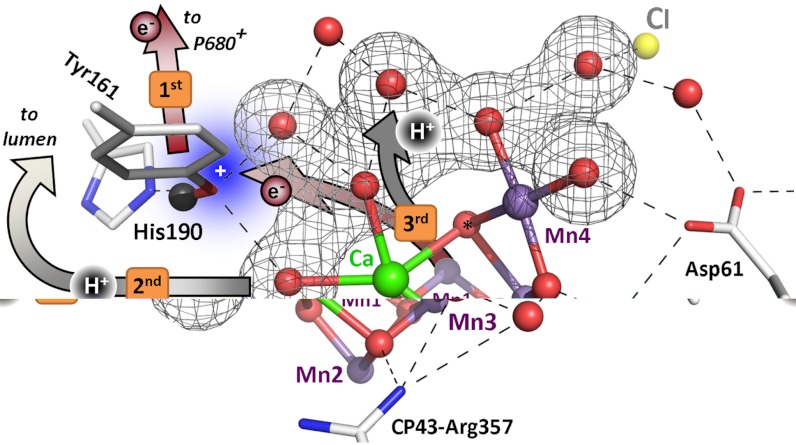
Fig. 5.
Sequence of events in the classical S2 → S3 transition of photosynthetic water oxidation. The Mn4CaO5 cluster, the redox-active tyrosine (Tyr161), and the key groups of the surrounding hydrogen-bonded network (15) are shown. All indicated amino acid residues are from the D1 subunit of PSII, with exception of CP43–Arg357. (Water molecules, HxO, are indicated as red spheres; putative H-bonds as broken lines that connect H-bond donor and acceptor. Of all the protons, only the phenolic proton is shown as a grey sphere.) The grey mesh outlines a water cluster that includes 4 HxO in the first coordination sphere of manganese (Mn4), as well as the calcium (Ca) and three second-sphere water molecules. Within less than 100 ns after absorption of a photon and oxidation of the primary chlorophyll donor of PSII (P680), Tyr161 (YZ) is oxidized by P680+ (“1st”). 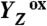 formation results in a rearrangement of the shown H-bonded network (completed within less than 1 µs), likely involving a shift of the phenolic proton to His190 and lowering of pK values for deprotonation of the water molecules in the outlined cluster (grey mesh). A proton is removed from the Mn complex/YZ environment within about 30 µs, as evidenced by the PBD results presented herein, and a proton vacancy is supposedly created within the outlined water cluster (“2nd”). In the ET to
formation results in a rearrangement of the shown H-bonded network (completed within less than 1 µs), likely involving a shift of the phenolic proton to His190 and lowering of pK values for deprotonation of the water molecules in the outlined cluster (grey mesh). A proton is removed from the Mn complex/YZ environment within about 30 µs, as evidenced by the PBD results presented herein, and a proton vacancy is supposedly created within the outlined water cluster (“2nd”). In the ET to 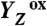 (about 300 µs), Mn oxidation is directly coupled to a proton transfer step involving the previously created proton vacancy of the water cluster (concerted electron–proton transfer) (“3rd”).
(about 300 µs), Mn oxidation is directly coupled to a proton transfer step involving the previously created proton vacancy of the water cluster (concerted electron–proton transfer) (“3rd”).