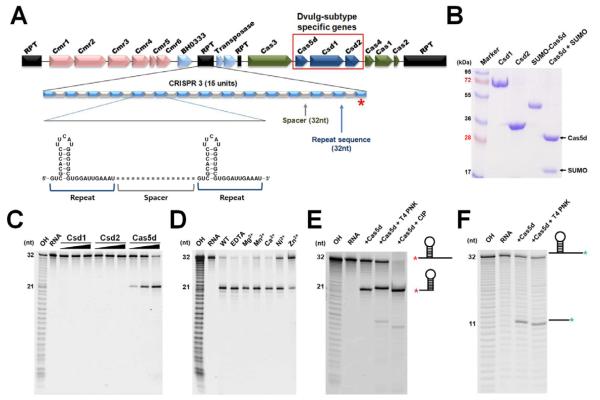
Figure 1. Cas5d is the pre-crRNA processor in the Subtype I-C/Dvulg CRISPR-Cas system.
(A) Diagram of the CRISPR/Cas system in B. halodurans C-125, which consists of three CRISPR loci, four core cas genes (cas1, cas2, cas3, and cas4), three Subtype I-C subtype specific cas genes (csd1, csd2 and cas5d), and a Subtype III-B cas genes. The organization and representative sequence of the CRISPR 3 locus is illustrated. (B) SDS-PAGE of the purified subtype-specific B. halodurans Csd1, Csd2 and Cas5d proteins. (C) Pre-crRNA processing assay identified processing activity in Cas5d, but not in Csd1 or Csd2. 5′-HEX-labeled CRISPR repeat RNA (0.2 μM) was used as the substrate with a titration of purified Csd1, Csd2, Cas5d, or equimolar amount of three proteins together (0.04, 0.2 and 1.0 μM). Cas5 cleaved between G21 and U22 in the 32-nt repeat sequence as indicated by the alkaline hydrolysis (OH) sequence ladder. (D) Metal ion-independent endoribonuclease activity of Cas5d. Cleavage reaction was carried out in the presence of 10 mM Mg2+, Mn2+, Ca2+, Ni2+, Zn2+, or EDTA. (E) To verify the presence of a 2′,3′-cyclic phosphate in the 5′-half of the crRNA product, 0.2 μM 5′-HEX were incubated with 1 μM Cas5d at 37 °C for 20 min, the reactions were then further incubated with either T4 polynucleotide kinase (PNK) or Calf Intestine Phosphotase (CIP) for an additional 30 min, separated on 18% sequencing gel, and scanned using typhoon 2900. T4 PNK, but not CIP, is capable of removing the 2′,3′-cyclic phosphate, causing the 5′ product to migrate slower. (F) To verify the presence of a 5′-OH in the 3′-half of the cleavage product, the 3′-Fluoresein labeled pre-crRNA repeats were incubated with Cas5d in similar reaction condition. Further incubation with T4 PNK in the presence of ATP added a phosphate to the 5′-OH and caused the 3′ product to migrate slightly faster. See also Figure S1.