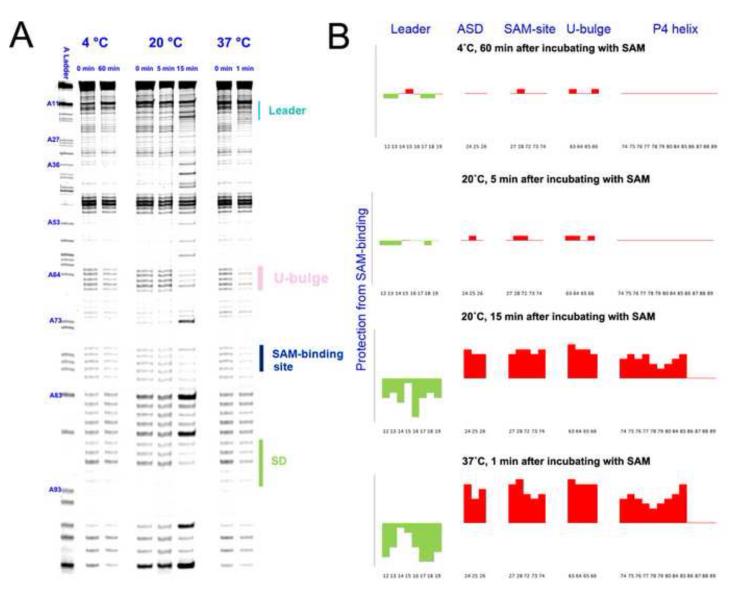
Figure 4.
Time-resolved SHAPE analysis revealed a steep temperature dependency in the conformational transition of the SMK box riboswitch. (A) Time-dependent SHAPE resolved on the sequencing gel. (Left) At 4 °C, SMK remained in the ON conformation after 60 minutes of SAM introduction. (Middle) At 20 °C, SMK mostly remained in the ON conformation after 5 minutes of SAM introduction, but switched to mostly OFF conformation after 15 minutes of incubation. (Right) At 37 °C, SMK completed the ON-to-OFF transition within a minute. Residue numbers were labeled next to the sequencing lane on the left. Important structural motifs were marked on the right. Temperatures and time points are marked on the top. (B) Quantified reactivity profile revealed structural changes upon SAM binding. Upward red bars represent protected residues due to SAM-induced structure formation. Green downward bars represent exposed bases upon SAM-binding. Residue numbers and corresponding structural motifs are indicated.