Abstract
Introduction
Androgen receptor [CAG]n microsatellite has been linked to human diseases.
Methods
Six nonhuman primates were genotyped for the [CAG]n microsatellite.
Results
Marmosets and macaques are monomorphic while mangabeys, baboons and chimpanzees are polymorphic.
Conclusions
Nonhuman primates that are polymorphic for the microsatellite are candidate animal models for CAG-related diseases.
Introduction
The androgen receptor (AR) gene is more than 90 kb long and is located on chromosome Xq11-12 in humans [6, 15]. The gene contains eight exons and encodes a protein with three functional domains: the N-terminal domain, a DNA-binding domain, and an androgen-binding domain [6, 13]. The androgen receptor helps direct the development of male sexual characteristics and regulate other important functions in both males and females. The AR gene contains two trinucleotide repeat segments in exon 1: [CAG]n codes for the polyglutamine tract (polyQ) while [GGN]n codes for the polyglycine tract (polyG) in the AR protein. In humans, the number of CAG repeats in the AR gene ranges from fewer than 10 to about 36 [4, 10,12]. Longer polyglutamine segments in humans lead to spinal bulbar muscular atrophy (SBMA) also called Kennedy’s disease [14]. Short polyglutamine tracts have been linked to increased risk [11] and early onset [9] of prostate cancer. A negative correlation has also been reported between CAG repeats and serum levels of prostate specific antigen (PSA) in subfertile men [16]. The androgen receptor has also been associated with coronary heart disease (CHD) [1, 19].
The aim of this study was to genotype AR CAG polymorphisms in nonhuman primate species used in biomedical research and then use this information to identify the best animal model for studying the relationship between AR CAG repeats, and prostatic diseases and CHD risk factors.
Materials and Methods
Animal tissues
DNA samples from 230 baboons (Papio hamadryas), 23 cynomolgus macaques (Macaca fascicularis), 54 rhesus macaques (Macaca mulatta), 56 sooty mangabeys (Cercocebus atys), 48 marmosets (Callithrix jacchus), and 48 chimpanzees (Pan troglodytes) were used. Only male animals were used.
Baboon, chimpanzee, marmoset, rhesus and cynomogus monkey samples were obtained from the Southwest National Primate Research Center (SNPRC), Texas Biomedical Research Institute, San Antonio. Sooty mangabey DNA samples were from Yerkes National Primate Research Center, Atlanta. Founders of the baboon captive population were wild-caught olive baboons (P. hamadryas anubis) and yellow baboons (P. h. cynocephalus) from East Africa (360 females and 40 males). The cynomolgus monkeys and chimpanzees are the progeny of many founder animals that were obtained from various sources. The marmosets are descendants of more than 40 animals obtained from several sources in Europe and the USA. The genetic background of the rhesus monkey and sooty mangabey colonies is not very diverse as they are descended from few founder animals.
[CAG]n microsatellite genotyping
DNA for the microsatellite assays was extracted from frozen tissues using the DNeasy kit (Qiagen). Polymerase chain reactions were run using Platinum Taq DNA polymerase (Invitrogen). The primer sequences used for CAG microsatellite amplification are located in AR exon 1(see below).
AR Forward: 5′-ACCGAGGAGCTTTCCAGA AT-3′
AR Reverse: 5′-GAAGGCTGCTGTTCCTCATC-3′
PCR products were separated on a 2% agarose gel, and bands were cut out from the gel and then purified using the Qiaquick Gel Extraction Kit (Qiagen). The Advanced Nucleic Acid Core Facility at the University of Texas Health Science Center at San Antonio performed the nucleotide sequencing.
Results
CAG repeat polymorphisms
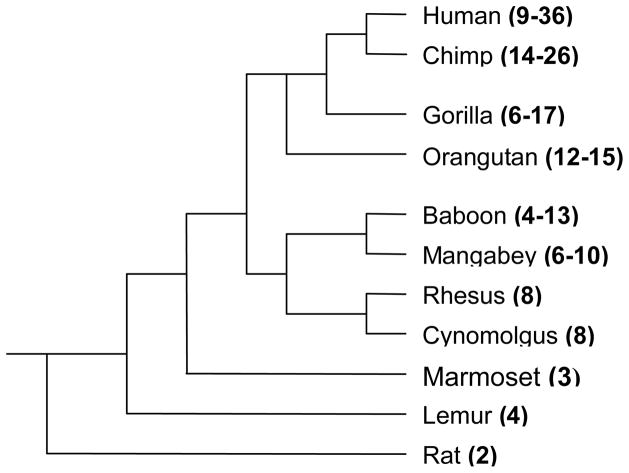
Three species (cynomolgus macaques and rhesus macaques, and marmosets) were found to be monomorphic. Only one genotype of 3 CAG repeats was found in marmosets and only one genotype of 8 CAG repeats was found in the macaques. Baboons, sooty mangabeys and chimpanzees, on the other hand, show a high level of polymorphism. In baboons there were 10 alleles ranging from 4 to 13 repeats, while in sooty mangabeys there were 3 genotypes ranging from 8 to 10. In chimpanzees alleles ranged from 14 to 26 repeats. The allele frequencies for each species studied are shown in Table 1. Using data from the present study combined with published data on CAG repeats from other primates, we have constructed a phylogenetic tree that show an increase in CAG microsatellite length as we move from species that are distantly related to humans towards those that are closely related to humans (Figure 1). A similar tree was published previously [3, 17].
Table 1.
Androgen receptor CAG genotypes in 6 male nonhuman primate species
| Baboon (n=230) | |
| CAG repeats | Frequency (%) |
| 4 | 0.87 |
| 5 | 3.04 |
| 6 | 6.52 |
| 7 | 4.78 |
| 8 | 19.57 |
| 9 | 41.30 |
| 10 | 16.52 |
| 11 | 6.52 |
| 12 | 0.43 |
| 13 | 0.43 |
| Chimpanzee (n=48) | |
| CAG repeats | Frequency (%) |
| 14 | 4.35 |
| 17 | 15.21 |
| 18 | 13.04 |
| 19 | 10.87 |
| 20 | 4.35 |
| 21 | 19.56 |
| 22 | 21.74 |
| 23 | 4.35 |
| 24 | 4.35 |
| 25 | 2.17 |
| 26 | 2.17 |
| Mangabey (n=56) | |
| CAG repeats | Frequency (%) |
| 8 | 69.64 |
| 9 | 3.57 |
| 10 | 26.79 |
| Cynomolgus monkey (n=23) | |
| CAG repeats | Frequency (%) |
| 8 | 100 |
| Rhesus monkey | (n=54) |
| CAG repeats | Frequency (%) |
| 8 | 100 |
| Marmoset (n=48) | |
| CAG repeats | Frequency (%) |
| 3 | 100 |
Figure 1.
Species differences in the number of CAG microsatellite repeats in the androgen receptor. The number of CAG repeats are in brackets. Rat and lemur CAG repeat numbers were based on data from Choong et al 1998. The human data was based on Huhtaniemi et al 2009, gorilla data was based on Djian et al 1996, the orangutan data was based on on GenBank sequence #AB207232 and the sequence available on University of California Santa Cruz genome browser gateway (chrX:65097837-65098089). A version of this figure was previously published in Perelman et al 2011.
Discussion
Of the six species we sequenced, the marmosets represent New World monkeys, the macaques, baboons and sooty mangabeys represent Old World monkeys and the chimpanzees are anthropoid apes. Previous studies have reported a linear increase in [CAG]n repeat length proportional to the time of species divergence from humans [3]. Our study builds upon those previous studies by both examining more samples of each species and including previously unstudied species to confirm the previous findings. The results reported here confirm that the increase in [CAG]n repeat length from (3 repeats) in marmosets to macaques (8 repeats), baboons (4–13 repeats) to chimpanzees (14–27 repeats) parallels their evolutionary relationships. One drawback of our study could be the lack of genetic diversity in the primate colonies studied, although this effect is limited as regards baboons, cynomolgus monkeys, chimpanzees and marmosets because they are the progeny of many founder animals. The absence of polymorphisms in the rhesus colony could be due to the fact that this colony is genetically close.
It is clear from this study that the normal physiological range for androgen receptor [CAG]n repeats in nonhuman primates is very variable. The [CAG]n microsatellite is in the coding region of the gene, but there is little understanding of its role. It seems that a considerable length of these [CAG]n repeats can be tolerated. This might suggest that the [CAG]n repeats may not always play a critical function but raises the question of why they are retained in all the species studied so far. One possibility is that the variation in [CAG]n repeats length represents a mechanism for fine-tuning the activity of this gene [18]. This could be plausible considering that the [CAG]n microsatellite is located in the N-terminal domain, which has been reported to be a part of the gene that influences transcription efficiency [2].
In humans the [CAG]n microsatellite is highly polymorphic. Longer repeats (≥40) have been reported to lead to Kennedy’s disease whereas shorter CAG repeats have been reported to result in higher activation of the receptor thus increasing the risk for several diseases [4, 7, 8].
In summary we have shown that the AR gene is polymorphic in baboons, sooty mangabeys and chimpanzees but monomorphic in macaques and marmosets. Nonhuman primates that are polymorphic for the CAG repeat could potentially be good models for investigation of the control of the AR gene activity, as well as human disease conditions that are associated with variation in this microsatellite.
Acknowledgments
We thank Dr. Suzette Tardif for providing the marmoset tissues, Dr. Zach Johnson at the Yerkes National Primate Research Center, for the sooty mangabey DNA samples and Roy E. Garcia IV for technical assistance.
Grant Sponsor:
This work was supported by the Voelcker Foundation grants to James N. Mubiru and by NIH grants P51 RR0139986 and K01RR025161-01 from the National Center for Research Resources.
This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant number C06 RR013556 from the National Center for Research Resources, National Institutes of Health.
Literature Cited
- 1.Alevizaki M, Cimponeriu AT, Garofallaki M, Sarika HL, Alevizaki CC, Papamichael C, Philippou G, Anastasiou EA, Lekakis JP, Mavrikakis M. The androgen receptor gene CAG polymorphism is associated with the severity of coronary artery disease in men. Clin Endocrinol (Oxf) 2003;59:749–55. doi: 10.1046/j.1365-2265.2003.01917.x. [DOI] [PubMed] [Google Scholar]
- 2.Barrack ER. Androgen receptor mutations in prostate cancer. Mt Sinai J Med. 1996;63:403–12. [PubMed] [Google Scholar]
- 3.Choong CS, Kemppainen JA, Wilson EM. Evolution of the primate androgen receptor: a structural basis for disease. J Mol Evol. 1998;47:334–42. doi: 10.1007/pl00006391. [DOI] [PubMed] [Google Scholar]
- 4.Choong CS, Kemppainen JA, Zhou ZX, Wilson EM. Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol Endocrinol. 1996;10:1527–35. doi: 10.1210/mend.10.12.8961263. [DOI] [PubMed] [Google Scholar]
- 5.Djian P, Hancock JM, Chana HS. Codon repeats in genes associated with human diseases: fewer repeats in the genes of nonhuman primates and nucleotide substitutions concentrated at the sites of reiteration. Proc Natl Acad Sci USA. 1996;93:417–21. doi: 10.1073/pnas.93.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Giovannucci E, Platz EA, Stampfer MJ, Chan A, Krithivas K, Kawachi I, Willett WC, Kantoff PW. The CAG repeat within the androgen receptor gene and benign prostatic hyperplasia. Urology. 1999;53:121–5. doi: 10.1016/s0090-4295(98)00468-3. [DOI] [PubMed] [Google Scholar]
- 8.Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA. 1997;94:3320–3. doi: 10.1073/pnas.94.7.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy DO, Scher HI, Bogenreider T, Sabbatini P, Zhang ZF, Nanus DM, Catterall JF. Androgen receptor CAG repeat lengths in prostate cancer: correlation with age of onset. J Clin Endocrinol Metab. 1996;81:4400–5. doi: 10.1210/jcem.81.12.8954049. [DOI] [PubMed] [Google Scholar]
- 10.Huhtaniemi IT, Pye SR, Limer KL, Thomson W, O’Neill TW, Platt H, Payne D, John SL, Jiang M, Boonen S, Borghs H, Vanderschueren D, Adams JE, Ward KA, Bartfai G, Casanueva F, Finn JD, Forti G, Giwercman A, Han TS, Kula K, Lean ME, Pendleton N, Punab M, Silman AJ, Wu FC European Male Ageing Study Group. Increased estrogen rather than decreased androgen action is associated with longer androgen receptor CAG repeats. J Clin Endocrinol Metab. 2009;94:277–84. doi: 10.1210/jc.2008-0848. [DOI] [PubMed] [Google Scholar]
- 11.Irvine RA, Yu MC, Ross RK, Coetzee GA. The CAG and GGC microsatellites of the androgen receptor gene are in linkage disequilibrium in men with prostate cancer. Cancer Res. 1995;55:1937–40. [PubMed] [Google Scholar]
- 12.Jin B, Beilin J, Zajac J, Handelsman DJ. Androgen receptor gene polymorphism and prostate zonal volumes in Australian and Chinese men. J Androl. 2000;21:91–8. [PubMed] [Google Scholar]
- 13.Kuiper GG, Faber PW, van Rooij HC, van der Korput JA, Ris-Stalpers C, Klaassen P, Trapman J, Brinkmann AO. Structural organization of the human androgen receptor gene. J Mol Endocrinol. 1989;2:R1–4. doi: 10.1677/jme.0.002r001. [DOI] [PubMed] [Google Scholar]
- 14.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–9. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 15.Lubahn DB, Joseph DR, Sar M, Tan J, Higgs HN, Larson RE, French FS, Wilson EM. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol. 1988;2:1265–75. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- 16.Mifsud A, Choon AT, Fang D, Yong EL. Prostate-specific antigen, testosterone, sex-hormone binding globulin and androgen receptor CAG repeat polymorphisms in subfertile and normal men. Mol Hum Reprod. 2001;7:1007–13. doi: 10.1093/molehr/7.11.1007. [DOI] [PubMed] [Google Scholar]
- 17.Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O’Brien SJ, Pecon-Slattery J. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. Epub 2011 Mar 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein DC, Leggo J, Coetzee GA, Irvine RA, Buckley M, Ferguson-Smith MA. Sequence variation and size ranges of CAG repeats in the Machado-Joseph disease, spinocerebellar ataxia type 1 and androgen receptor genes. Hum Mol Genet. 1995;4:1585–90. doi: 10.1093/hmg/4.9.1585. [DOI] [PubMed] [Google Scholar]
- 19.Zitzmann M, Brune M, Kornmann B, Gromoll J, von Eckardstein S, von Eckardstein A, Nieschlag E. The CAG repeat polymorphism in the AR gene affects high density lipoprotein cholesterol and arterial vasoreactivity. J Clin Endocrinol Metab. 2001;86:4867–73. doi: 10.1210/jcem.86.10.7889. [DOI] [PubMed] [Google Scholar]