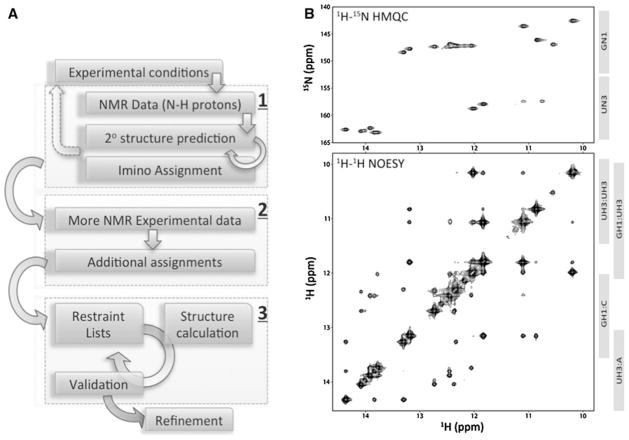
Fig. 1.
a Standard steps for solving RNA structures using NMR. The process can be conceptually divided into three main steps: 1 imino proton assignments and secondary structure validation, 2 full resonance assignment and restraint list construction, 3 structure calculation. The vast majority of work in these steps requires manual intervention—although some automation support is available for structure calculation and refinement. Circular arrows indicate some of the possible steps for iterative refinement while the dotted arrow suggests the potential need for construct modifications. Imino regions of 2D NOESY and HMQC NMR spectra are often highly informative for step 1. b Two types of NMR spectra containing information relevant for assigning NMR signals for imino protons and nitrogens in an RNA molecule (BMRB ID 17921)