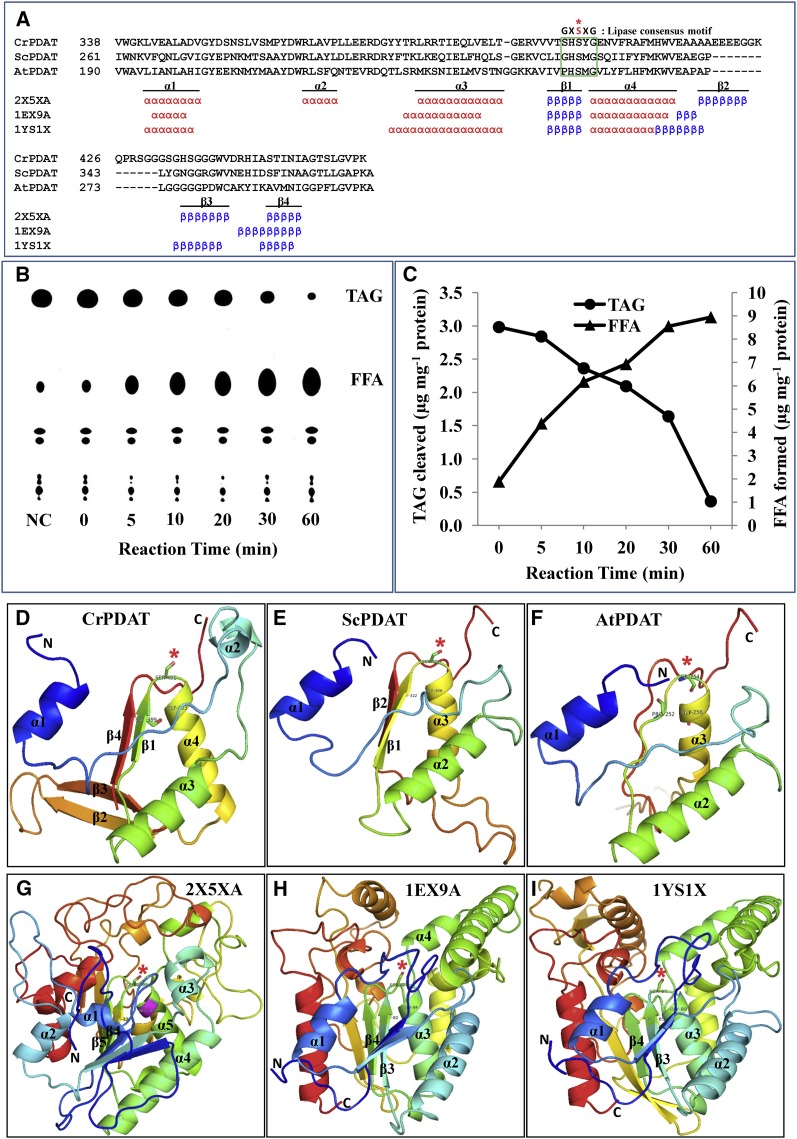
Figure 4.
Cr-PDAT Possesses a Lipase Functional Domain and TAG Lipase Activity.
(A) Sequence alignment and secondary structure elements between the PDAT-like proteins and their referenced templates within α/β hydrolase fold region. The conserved lipase motif is indicated by the green box. The essential Ser (S) residue that constitutes the catalytic triad for the active site is shown as a red asterisk.
(B) TAG lipase activity was detected by monitoring the formation of free fatty acids (18:1) and the degradation of TAG at various time points. Heat-inactivated PDAT was used as negative control (NC); reaction time for the negative control was 60 min. FFA, free fatty acid.
(C) Quantitative analysis of TAG and fatty acids in the enzymatic assay.
(D) to (F) Comparison of the predicted structure of conserved α/β hydrolase fold domains of Cr-PDAT, Sc-PDAT, and At-PDAT, respectively.
(G) to (I) Templates used for construction of structures of Cr-PDAT, Sc-PDAT, and At-PDAT, respectively. The rainbow color code describes the three-dimensional structures from the N (blue) to C termini (red); specific α-helices (α) and β -sheets (β) were identified as well as the active site Ser (asterisk).