Figure 3.
Siderophore Biosynthesis Is Upregulated in the ΔhapX Mutant during Iron Starvation.
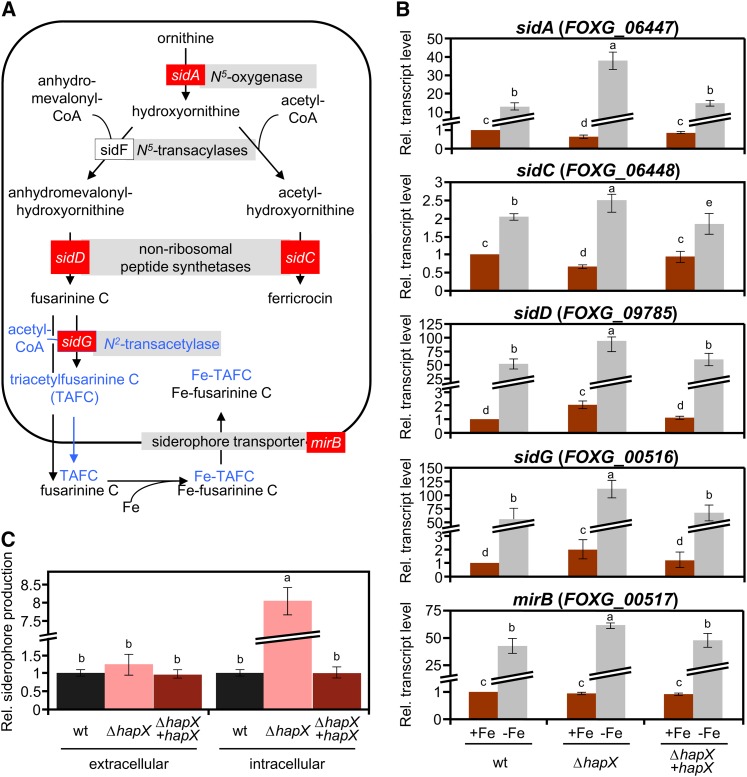
(A) Siderophore biosynthetic pathways based on the information available in Aspergillus. Genes marked in red were used for transcriptional analysis in F. oxysporum (B). Putative conversion of FsC to TAFC suggested by genome inspection, but not detected in the siderophore analysis, is shown in blue.
(B) Quantitative real-time RT-PCR analysis of the indicted genes was performed in fungal strains grown as described in Figure 2. Transcript levels of sidA, sidC, sidD, sidG, and mirB genes are expressed relative to those of the wild-type (wt) strain grown under iron-replete conditions. Bars represent se from three independent experiments with three technical replicates each.
(C) CAS assay-mediated quantification of extra- and intracellular siderophore production during iron starvation, normalized to the wild-type strain. Bars represent se from three independent experiments with three technical replicates each. Values with the same letter are not significantly different according to the Mann-Whitney test (P = 0.05).
[See online article for color version of this figure.]