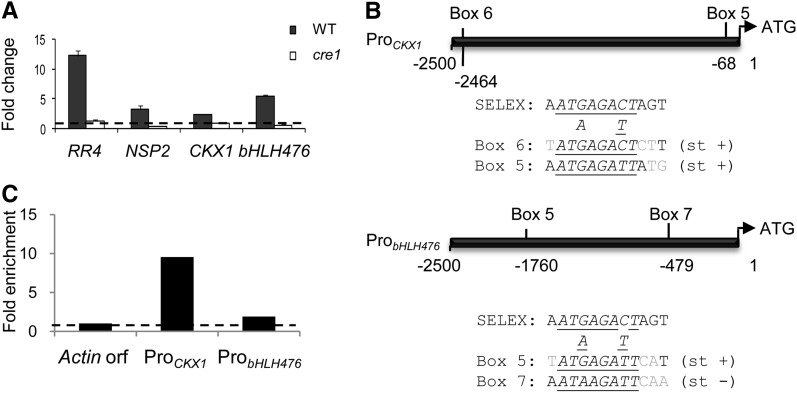
Figure 4.
CRE1-Dependent Regulation of Primary Cytokinin Response Genes.
(A) Real-time RT-PCR analysis of selected candidate cytokinin primary response genes in roots of the wild type (WT) or of a CRE1 cytokinin receptor mutant (cre1-1) treated for 1 h with 10−7 M of BAP. Genes tested are RR4, NSP2, CKX1, and bHLH476. The nontreated condition was set to 1 (dotted line) to visualize FCs. Error bars represent sd of two technical replicates, and one representative biological replicate out of four is shown (n > 10 independent transgenic roots/condition).
(B) Schematic diagram of the CKX1 and bHLH476 promoters. The different RRBS cis-elements variants (boxes 5 to 7) are indicated. Alignment to the major SELEX consensus is shown below each promoter diagram. Gray indicates nonconserved nucleotides, underlined indicates conserved nucleotides between all boxes of each promoter, italics indicates 8-bp RRBS core, and st + or st − indicates DNA strand of sequences shown.
(C) ChIP-qPCR of RR1 binding to CKX1 and bHLH476 promoters. Actin11 ORF was used as a negative control, and IP values were normalized for each gene against the input genomic DNA. Ratios with Actin11 were calculated to visualize RR1 binding fold enrichment (n > 30 independent transgenic roots/construct).