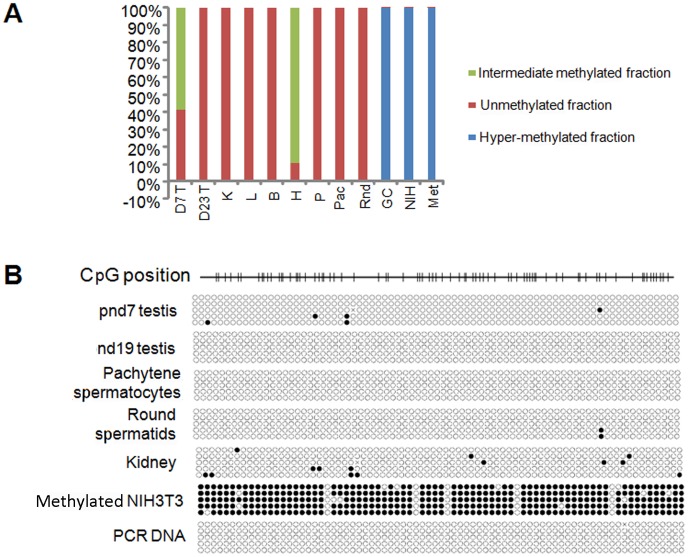
Figure 7. Tissue-specific Ccna1 expression is not modulated by differential promoter methylation.
(A) Gross methylation analysis of the Ccna1 promoter in tissues and cell lines as follows: mouse testis (T) at the indicated age, kidney (K), liver (L), brain (B), heart (H), pancreas (P), purified pachytene cells (Pac), round spermatids (Rnd) and cell lines GC-4spc (GC), NIH3T3 (NIH). For each analysis, 500 ng of total genomic DNA was divided into four and assigned to mock control, methylation-sensitive, methylation-dependent and double digestion respectively. After digestion with methylation sensitive and insensitive nucleases, the methylation profile was obtained from the Ct values of real-time PCR with Ccna1-CpG methylation specific primers. (B) The methylation status of individual CpG residues on the Ccna1 promoter was analyzed by bisulfite sequencing. One µg of DNA from the indicated samples was subjected to bisulfite conversion followed by amplification with primers specific for the Ccna1 CpG island. PCR products were cloned into a pGEMTeasy vector, sequenced, and sequences were analyzed with QUMA software. In vitro CpG methylated NIH3T3 DNA was used as a positive control and a PCR amplified DNA from unconverted sequence was used as a negative control. Non-methylated CpG are indicated as open circles. The Ccna1 promoter with CpG positions is depicted schematically.