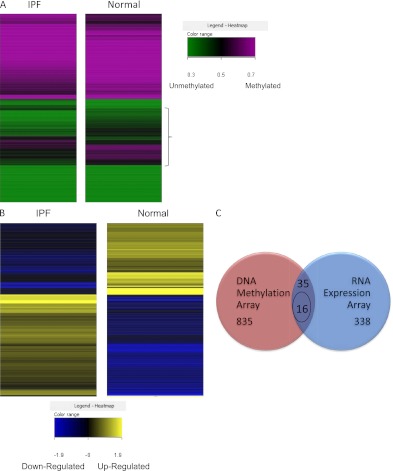
Figure 2.
(A) Heatmap of DNA methylation array: methylated/unmethylated genes in idiopathic pulmonary fibrosis (IPF) and normal control samples. Each row represents a gene with its average methylation status of IPF samples or normal samples. A gradient color scale ranging between green (unmethylated) and purple (methylated) is included. Brackets indicate genes with the most significant differences in methylation status. (B) Heatmap of gene expression array in IPF and normal samples. Each row represents a gene with its average expression of IPF samples or normal samples. A gradient color scale ranging between blue (down-regulated in RNA expression) and gold (up-regulated in RNA expression) is shown. (C) Venn diagram of overlapped genes from DNA methylation array and RNA gene expression array data sets. Thirty-five genes in both data sets have a |DiffScore| of at least 13, which is equivalent to a P value less than 0.05, and are more than twofold changed in the RNA array, and 16 of these genes have inverse DNA methylation and RNA expression (i.e., DNA hypermethylation with RNA down-regulation, or DNA hypomethylation with RNA up-regulation). Entity List 1 (red): IPF filtered on expression (0.0–100.0)th percentile in the raw data (translated from IPF methylation), 870 entities (835 + 35). Entity List 2 (blue): t test unpaired [IPF] versus [normal] p ≤ 0.05, fold change ≥ 2.0, 373 entities (338 + 35).