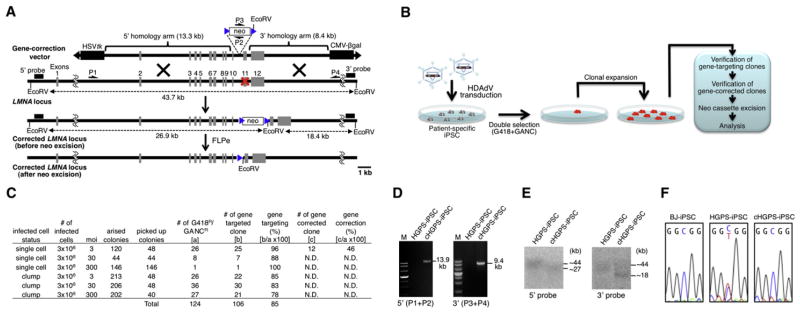
Figure 1. Correction of HGPS-Associated LMNA Mutation in iPSCs with LMNA-c-HDAdV.
(A) Schematic molecular representation of LMNA gene correction with LMNA-c-HDAdV. The primers for PCR are shown as arrows (P1, P2, P3, and P4). The probes for Southern analyses are shown as black bars (5′ probe and 3′ probe). HSVtk stands for herpes simplex virus thymidine kinase gene cassette used for negative selection; neo stands for neomycin-resistant gene cassette used for positive selection; CMV-β-gal indicates the β-gal expression cassette for determination of HDAdV titer; blue triangle, FRT site; red X, mutation sites in exon 11.
(B) Schematic representation of the gene-correction approach employed in iPSCs with laminopathy-associated mutation(s).
(C) Gene-targeting and gene-correction efficiencies at the LMNA locus in HGPS-iPSCs achieved by different infection conditions. N.D., not determined.
(D) PCR analyses of HGPS-iPSCs and cHGPS-iPSCs via 5′ primer pair (P1 and P2; 13.9 kb) or 3′ primer pair (P3 and P4; 9.4 kb). M, DNA ladder.
(E) Southern blot analyses of HGPS-iPSCs and cHGPS-iPSCs. The approximate molecular weights (kb) corresponding to the bands are indicated.
(F) Sequencing results of C1824T mutation site in exon 11 of LMNA in BJ-iPSCs (wild-type), HGPS-iPSCs, and cHGPS-iPSCs. All iPSCs employed represent high-passage iPSCs (>30).
See also Figure S1.