Figure 2.
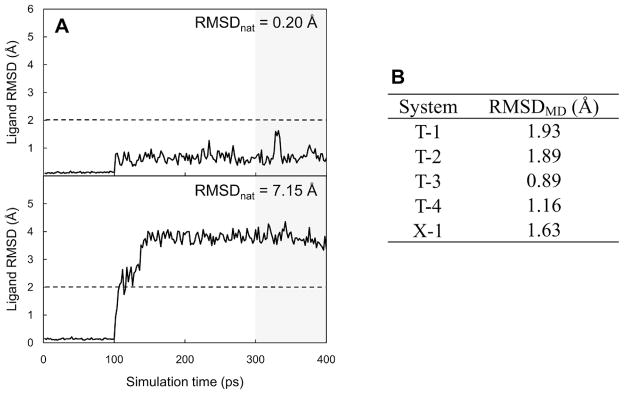
Structural stability of a ligand bound to a receptor during equilibration MD simulations. (A) Representative plots of ligand RMSD changes from an initial docked pose as a function of simulations time for near-native pose (RMSD from native pose, RMSDnat = 0.20 Å) and incorrectly docked pose (RMSDnat = 7.15 Å). These plots were prepared from the simulation results for T-3 cognate-receptor docking models. (B) Average ligand RMSDs from the native conformations during last 100-ps simulations (RMSDMD) for the crystal structures.
