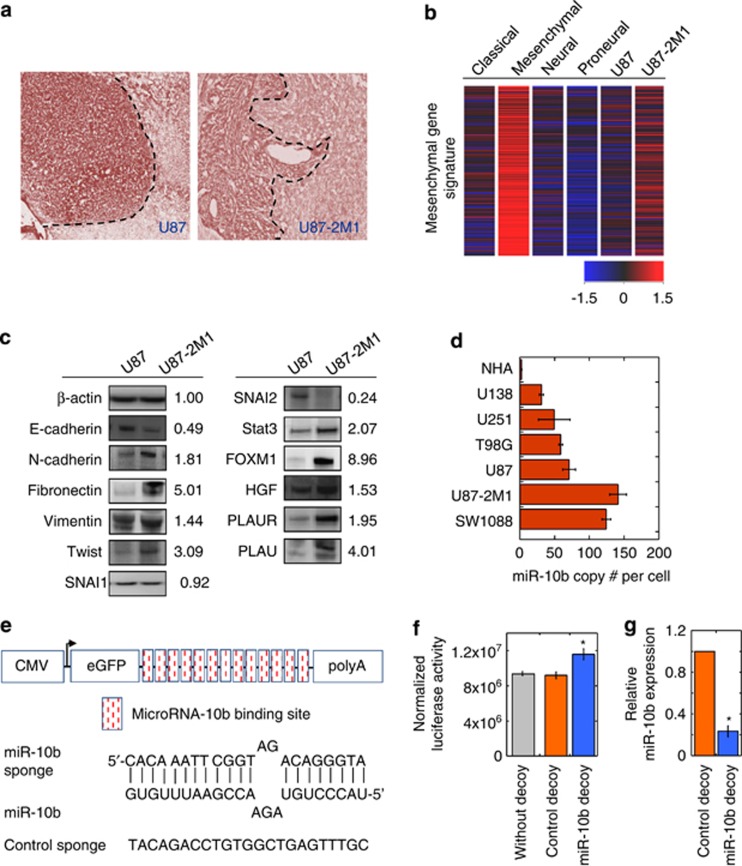
Figure 1.
Expression of miR-10b is elevated in invasive mesenchymal subtype-like U87-2M1 glioma cells. (a) Invasive U87-2M1 glioma cell growth in the brain. U87 and U87-2M1 glioma cells were grown intracranially in nude mice for 3 weeks. Tumor growth was visualized by H&E staining of brain tissues. Bars equal 200 μm. (b) Heatmap distinctly shows that the mesenchymal gene expression signature is highly activated in the mesenchymal subtype of GBM and U87-2M1 cells, but not in the neural, proneural and classical subtypes, of GBM and U87 glioma cells. (c) Western blotting analysis of expression of invasiveness-associated proteins in U87-2M1 cells as compared with those in U87 cells. Numbers indicate relative protein expression in U87-2M1 compared with U87 cells after normalization to beta-actin levels. Blots shown are representative of biological triplicates. (d) qPCR quantification of miR-10b expression in glioma cell lines (SW1088, U87-2M1, U87, T98G, U251 and U138) and NHA revealed an upregulation of miR-10b expression in glioma cell lines. (e) Construction of control or miR-10b decoy vector. Upper, schematic of the expression cassette. Bottom, sequence of the miR-10b-binding site and control sequence with no known complementarity to any known miRNA. (f) miR-10b decoy vector, but not the control decoy vector, relieves the suppression of luciferase expression by endogenous miR-10b in U87-2M1 glioma cells. Error bars represent S.E.from three independent experiments; *P<0.05. (g) miR-10b decoy vector decreases the level of detectable miR-10b in U87-2M1 glioma cells. Error bars represent S.E. from three independent transductions; *P<0.05