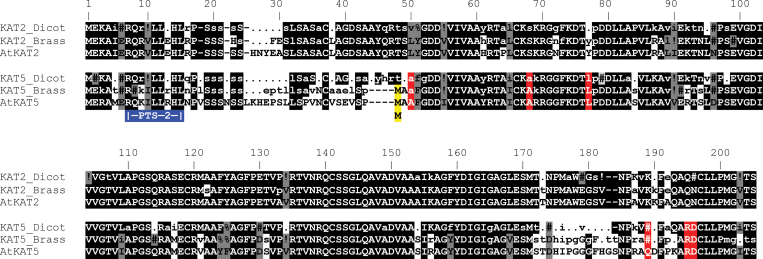
Fig. 2.
Alignment of dicotyledonous plant thiolase proteins. Consensus sequences were derived from four groups of sequences comprising dicotyledonous KAT2 or KAT5 and Brassicales KAT2 or KAT5 sequences. AtKAT2 and AtKAT5 are included as samples of specific sequences. Typical plant KAT proteins are about 460 amino acids; the first approximately 200 residues are shown. Highly conserved residues (present in >90% of sequences) in each of the four consensus sequences are shown in capital letters, while moderately conserved residues (present in 50–90% of sequences) in lower case. Non-conserved positions are depicted by a period (.) and gaps introduced into the alignment by a dash (-). Symbols used to indicate residues with strongly similar properties on the Gonnet PAM 250 matrix are: # (N, D, Q or E); % (F or Y);! (I or V). Residues conserved between the different consensus sequences are highlighted with black shading and grey shading indicates consensus sequence residues that are not identical but have similar properties. The red highlighting indicates residues that clearly distinguish KAT5. The blue box shows the PTS2 near the N-terminus of the proteins and the yellow highlighting the alternative initial Met that is conserved in all Brassicales KAT5 proteins.