Background: The sigma-1 receptor modulates the activity of ion channels.
Results: Atomic force microscopy imaging of complexes between sigma-1 receptors and Nav1.5 Na+ channels reveals a 4-fold symmetry.
Conclusion: Each of the four sets of six transmembrane regions in Nav1.5 constitutes a sigma-1 receptor binding site.
Significance: The sigma-1 receptor likely interacts with the transmembrane regions of its protein partners.
Keywords: Atomic Force Microscopy, Cancer, Ion Channels, Membrane Proteins, Molecular Imaging, Protein-Protein Interactions, Receptors, Sigma Receptor, Sodium Channels
Abstract
The sigma-1 receptor (Sig1R) is up-regulated in many human tumors and plays a role in the control of cancer cell proliferation and invasiveness. At the molecular level, the Sig1R modulates the activity of various ion channels, apparently through a direct interaction. We have previously shown using atomic force microscopy imaging that the Sig1R binds to the trimeric acid-sensing ion channel 1A with 3-fold symmetry. Here, we investigated the interaction between the Sig1R and the Nav1.5 voltage-gated Na+ channel, which has also been implicated in promoting the invasiveness of cancer cells. We show that the Sig1R and Nav1.5 can be co-isolated from co-transfected cells, consistent with an intimate association between the two proteins. Atomic force microscopy imaging of the co-isolated proteins revealed complexes in which Nav1.5 was decorated by Sig1Rs. Frequency distributions of angles between pairs of bound Sig1Rs had two peaks, at ∼90° and ∼180°, and the 90° peak was about twice the size of the 180° peak. These results demonstrate that the Sig1R binds to Nav1.5 with 4-fold symmetry. Hence, each set of six transmembrane regions in Nav1.5 likely constitutes a Sig1R binding site, suggesting that the Sig1R interacts with the transmembrane regions of its partners. Interestingly, two known Sig1R ligands, haloperidol and (+)-pentazocine, disrupted the Nav1.5/Sig1R interaction both in vitro and in living cells. Finally, we show that endogenously expressed Sig1R and Nav1.5 also functionally interact.
Introduction
The sigma-1 receptor (Sig1R)3 is widely expressed in both the central nervous system and peripheral tissues (1, 2), and a variety of functions have been ascribed to it, including modulation of voltage-gated (3–11), ligand-gated (12–15), volume-regulated (16) and acid-sensitive (17) ion channel activity at the plasma membrane, control of Ca2+ release from the endoplasmic reticulum (18), and neuroprotection in cerebral ischemia and stroke (19). Of most relevance to the present study, the Sig1R is known to be up-regulated in many human tumors and has been implicated in the control of cancer cell proliferation, resistance to apoptosis, and invasiveness (16, 20, 21).
The Sig1R shares 30% identity and 67% similarity with a yeast sterol C8-C7 isomerase (ERG2; 22). The receptor contains two transmembrane regions, although it is still unclear whether the N and C termini are cytoplasmic (5) or extracytoplasmic (18). The Sig1R is modulated by a wide variety of ligands (1, 2), including antipsychotic drugs (e.g. haloperidol) and psychotomimetics (e.g. pentazocine). Indeed, most of the evidence for the physiological relevance of the Sig1R has relied on the effects of these ligands; in contrast, there have been relatively few reports of functional effects of ligand-free Sig1Rs (e.g. Refs. 5, 11, 16, 18). Evidence has been presented recently that the hallucinogen N,N-dimethyltryptamine is an endogenous ligand at the Sig1R (23).
There is some evidence that modulation of ion channel function by the Sig1R is direct; for instance, its effects on the Kv1.4 voltage-gated K+ channel do not involve transduction mechanisms such as G protein signaling or phosphorylation (24). Furthermore, the Sig1R can be co-immunoprecipitated with Kv1.4 from membrane lysates prepared from both rat posterior pituitary cells and mRNA-injected Xenopus oocytes (5), and with the human ether-à-go-go-related gene (hERG) potassium channel from hERG/Sig1R-transfected human embryonic kidney (HEK)-293 cells (11). Using atomic force microscopy (AFM) imaging, we recently demonstrated at the single molecule level that the Sig1R directly interacts with the acid-sensing ion channel (ASIC)1a, which is known to assemble as a trimer (25, 26), forming a complex with 3-fold symmetry (27). This interaction likely underlies the ability of the Sig1R endogenously expressed in rat cortical neurons to inhibit ASIC1a-mediated membrane currents and thereby reduce consequent intracellular Ca2+ accumulation (17).
In the present study, we focused on the interaction between the Sig1R and the voltage-gated Na+ channel Nav1.5, which incorporates four sets of six transmembrane regions (TMRs) within a single polypeptide (28). Nav1.5 is responsible for the rapid upstroke of the action potential in cardiac myocytes and for the rapid propagation of the cardiac action potential. Significantly, it is also implicated in promoting the invasiveness of breast cancer cell lines such as MDA-MB-231 (29–31) and is known to be modulated by Sig1R ligands (3, 4, 23). For example, (+)-pentazocine reversibly inhibited Nav1.5 channels stably expressed in HEK-293 cells and in cardiac myocytes but had a much smaller effect in Sig1R knock-out myocytes (3). N,N-Dimethyltryptamine, the putative endogenous Sig1R ligand, had similar inhibitory effects on Nav1.5 currents and induced hypermobility in mice that was abrogated when the Sig1R was knocked out (23). In contrast, progesterone acted as an antagonist at the Sig1R, blocking the inhibitory effects of ligands such as N,N-dimethyltryptamine on Nav1.5 (4).
We set out to examine the structure of the complex formed between the Sig1R and Nav1.5 and to test whether the Sig1R/Nav1.5 interaction has functional consequences for Nav1.5 activity in MDA-MB-231 breast cancer cells. We show using AFM imaging that the Sig1R binds to Nav1.5 with 4-fold symmetry, suggesting that the Sig1R interacts with its partner proteins via their TMRs. We also show that Nav1.5 currents in MDA-MB-231 cells fall when Sig1R expression is reduced.
EXPERIMENTAL PROCEDURES
Cell Culture
tsA 201 cells (a subclone of HEK-293 cells stably expressing the SV40 large T-antigen) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, in an atmosphere of 5% CO2 and 95% air. The breast cancer cell line, MDA-MB-231, kindly provided by Dr. Laurent Combettes (Université de Paris-Sud), was cultured in DMEM supplemented with 10% fetal bovine serum and 50 units/ml penicillin and 50 μg/ml streptomycin.
Constructs
cDNA encoding the human Sig1R, with a C-terminal FLAG epitope tag, was subcloned into the vector pcDNA3.1/V5-His using HindIII and AgeI so as to delete the V5 epitope tag but leave the His6 tag. (The His6 tag was not used in any of the experiments described here.) The sequence of the construct was verified before use. cDNA encoding human Nav1.5, with a C-terminal hemagglutinin (HA) epitope tag, in the vector pcDNA3N, was kindly provided by Dr. C. Valdivia (University of Michigan).
Transient Transfection of tsA 201 Cells
Transient transfections of tsA 201 cells with DNA were carried out using the calcium phosphate precipitation method. A total of 250 μg of DNA was used to transfect cells in 5 × 162 cm2 culture flasks. For co-transfections with Sig1R-FLAG and Nav1.5-HA, 125 μg of DNA for each construct was used. After transfection, cells were incubated for 48 h at 37 °C to allow protein expression. Protein expression and intracellular localization were checked using immunofluorescence analysis of small-scale cultures. Cells were fixed, permeabilized, and incubated with appropriate primary antibodies (mouse monoclonal anti-FLAG (Sigma), mouse monoclonal anti-HA (Covance), rabbit polyclonal anti-HA (Sigma), and mouse monoclonal anti-Myc (Invitrogen), as a negative control), followed by either Cy3-conjugated goat anti-mouse, Cy3-conjugated goat anti-rabbit or fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibodies (Sigma). Cells were imaged by confocal laser scanning microscopy. To test for endogenous expression of the Sig1R, cell extracts were immunoblotted using a rabbit anti-Sig1R polyclonal antibody (Abcam). Immunoreactive bands were visualized using a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody followed by enhanced chemiluminescence.
Solubilization and Purification of Epitope-tagged Proteins
Transfected cells were solubilized in 1% Triton X-100 for 1 h, before centrifugation at 61,740 × g to remove insoluble material. To capture Nav1.5-HA, the solubilized extract was incubated with anti-HA-agarose beads (Sigma) for 3 h. The beads were washed extensively, and bound proteins were eluted with HA peptide (100 μg/ml). Sig1R-FLAG was captured in the same way using anti-FLAG-agarose beads and a triple-FLAG peptide (Sigma). Samples were analyzed by SDS-PAGE, followed by Coomassie blue staining and/or immunoblotting, using mouse monoclonal antibodies against HA or FLAG. Where appropriate, band densitometry was carried out using NIH ImageJ software (http://rsbweb.nih.gov/ij/).
Isolation of Biotinylated Proteins
To establish whether Sig1R-FLAG and Nav1.5-HA interacted at the plasma membrane, intact co-transfected cells were biotinylated by incubation with sulfo-NHS-LC-biotin (Pierce; 1 mg/ml) for 30 min. A detergent extract of the cells was then produced as described above. Biotinylated proteins were incubated with monomeric avidin-agarose (Pierce) for 1 h. The beads were washed extensively, and bound proteins were eluted with free biotin (2 mm). Eluted Sig1R-FLAG was captured using anti-FLAG-agarose beads and eluted with triple-FLAG peptide (Sigma). Samples were analyzed by SDS-PAGE, followed by immunoblotting, using mouse monoclonal antibodies against HA and FLAG.
AFM Imaging of Isolated Proteins
Isolated protein samples were diluted to a final concentration of 0.04 nm, and 45 μl of the sample was allowed to adsorb to freshly cleaved mica disks. After a 5-min incubation, the sample was washed with Biotechnology Performance Certified-grade water (Sigma) and dried under nitrogen. Imaging was performed with a Veeco Digital Instruments Multimode AFM controlled by a Nanoscope IIIa controller. Samples were imaged in air, using the tapping mode. The silicon cantilevers used had a drive frequency ∼300 kHz and a specified spring constant of 40 N/m (Olympus). The applied imaging force was kept as low as possible (As/A0 ∼0.85).
For individual Sig1R particles, molecular volumes were determined using Scanning Probe Image Processor (version 5; Image Metrology). It is well known that the geometry of the scanning AFM probe introduces a tendency to overestimate particle diameter. To minimize this probe convolution error, we used a Scanning Probe Image Processor particle threshold of 0.1 nm to provide accurate measurements of diameter. For particles within complexes, particle heights and diameters were measured manually using the Nanoscope software and used to calculate molecular volumes, according to Equation 1,
where h is the particle height and r is the radius (32). This equation assumes that the adsorbed particles adopt the form of a spherical cap.
Molecular volume based on molecular mass was calculated using Equation 2,
where M0 is the molecular mass, N0 is Avogadro's number, V1 and V2 are the partial specific volumes of particle (0.74 cm3/g) and water (1 cm3/g), respectively, and d is the extent of protein hydration (taken as 0.4 g water/g protein).
Selection of Binding Events
Several criteria were used to identify Nav1.5·Sig1R complexes. Heights and radii were measured for all particles, and the particle volumes were calculated. Bound particles needed to have a molecular volume between 30 and 120 nm3, which was the experimentally determined volume range for a Sig1R. A cross-section was drawn through the junction between the Sig1R and the adjacent Nav1.5 channel, and the height of the lowest point between receptor and channel was measured. This height needed to be greater than 0.3 nm for the Sig1R to be considered bound. Any particle was rejected if its length was greater than twice its width. To be considered a double binding event, all particles and both binding events needed to meet all of the above criteria.
Note that it has been shown previously (32) that the molecular volumes of proteins measured by imaging in air are similar to the values obtained by imaging under fluid; hence, the process of drying does not significantly affect the measured molecular volume. It has also been shown by the authors (33) and by others (32) that there is a close correspondence between the measured and predicted molecular volumes for various proteins over a wide range of molecular masses; hence, molecular volume is measured reasonably accurately by AFM imaging.
Statistical Analysis
Histograms were drawn with bin widths chosen according to Scott's equation,
where σ is an estimate of the S.D. and n is the sample size (34). Where Gaussian curves were fitted to the data, the number of curves was chosen so as to maximize the r2 value while giving significantly different means using Welch's t test for unequal sample sizes and unequal variances (35).
In Situ Proximity Ligation Assay
Cells growing on lysine- and collagen-coated glass coverslips in 3.5-cm diameter culture wells were co-transfected with 1.5 μg each of DNA encoding Nav1.5-HA and Sig1R-FLAG. Cells were incubated for 24 h at 37 °C to allow protein expression. Cells were fixed, permeabilized, and incubated with primary antibodies (rabbit polyclonal anti-HA plus mouse monoclonal anti-FLAG, both diluted 1:50) for a further 1 h. Cells were washed with phosphate-buffered saline and then incubated for 1 h with anti-mouse (+) and anti-rabbit (−) proximity ligation secondary antibodies, diluted 1:5, at 37 °C (36). These antibodies were obtained as part of a kit from Olink Bioscience, which also included ligation and amplification buffers. Cells were washed with phosphate-buffered saline and incubated in T4 DNA ligase diluted 1:40 in ligation buffer for 30 min at 37 °C. Cells were washed with TBS-T (150 mm NaCl, 0.05% Tween 20, 10 mm Tris, pH 7.4), and DNA amplification was performed by incubation with 1:80 DNA polymerase in amplification buffer for 100 min at 37 °C. This buffer also contained a fluorescent detection probe (excitation wavelength, 554 nm; emission wavelength, 576 nm). Cells were washed with 1× TBS (100 mm NaCl, 20 mm Tris, pH 7.5) and then 0.01× TBS. Coverslips were mounted on slides and imaged by confocal laser scanning microscopy.
Small Hairpin (sh)RNA Transduction of MDA-MB-231 Cells
The shRNA sequences targeted either to the Sig1R (GACTTCCTCACCCTCTTCTATCTCGAGATAGAAGAGGGTGAGGAAGTC) or to a random sequence (CAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGT) were subcloned into the mammalian expression vector pPRIpu as described previously (37) to generate the constructs pPRIGpu-shSig1R and pPRIGpu-shRD, respectively. Highly purified recombinant plasmids were obtained by anion-exchange chromatography (NucleobondAx, Macherey-Nagel). To generate retroviruses, HEK-293T cells were transfected the following day with 10 μg of either pPRIGpu-shSig1R or pPRIGpu-shRD, together with 5 μg of plasmid pCMV-VSV G and 5 μg of plasmid pCMV-gag-pol, using the calcium phosphate precipitation method. Six hours after transfection, cells were washed, and fresh medium was added. Replication-defective retroviruses were recovered in the culture medium between 24 h and 72 h post-transfection. For transduction experiments, MDA-MB-231 cells were seeded at 30–40% density in 100-mm dishes in culture medium. Retroviral supernatants were filtered through sterile 0.45-μm filters and then added directly to MDA-MB-231 cells in the presence of 4 μg/ml hexadimethrin bromide to enhance retroviral transduction efficiency. On day 6, puromycin (4 μg/ml) was added in fresh medium to begin selection of transduced cells.
Electrophysiology Experiments
MDA-MB-231 cells were prepared as previously described (29). The physiological saline solution used contained 5 mm KCl, 140 mm NaCl, 2 mm MgCl2, 1 mm CaCl2 and 10 mm HEPES, pH 7.4. The pipette solution contained 130 mm potassium aspartate, 2 mm MgCl2, 1 mm CaCl2, 10 mm EGTA, 2 mm ATP, 10 mm HEPES, pH 7.2. Soft glass patch electrodes (Brand, Wertheim, Germany) were prepared on a horizontal pipette puller (P-97, Sutter Instrument Co., Novato, CA) to achieve a final resistance of 2.5–4.0 megohms. Currents were recorded in whole cell configuration and voltage clamp mode at room temperature using an Axopatch 200B patch clamp amplifier (Axon instruments) with a DIGIDATA 1440 interface and pClamp10.2 software (Axon instruments). Analog signals were sampled at 10 kHz and filtered at 2 kHz. Cell capacitance and series resistance were electronically compensated by ∼60%. The P/5 subpulse correction of cell leakage was used to study the Na+ current. Na+ current-voltage (INa-V) relationships were constructed as described previously (29). Briefly, from a holding potential of −100 mV the membrane was stepped to potentials between −90 mV and +60 mV, in 5-mV increments, for 50 ms at a frequency of 2 Hz.
RESULTS
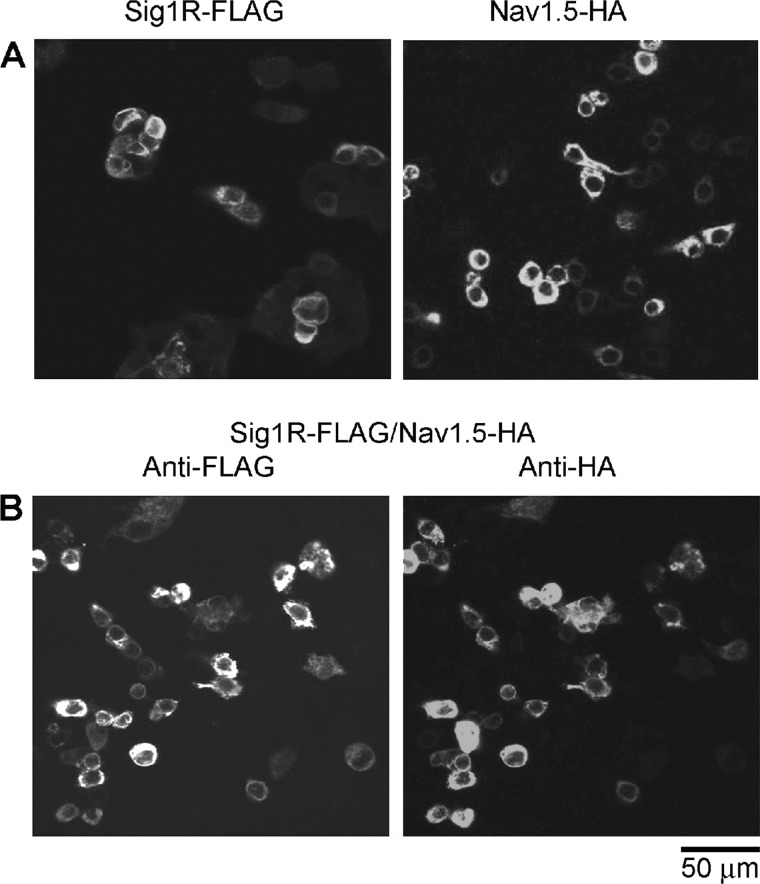
tsA 201 cells were transfected with DNA encoding either Sig1R-FLAG, Nav1.5-HA, or both. Protein expression and localization was confirmed by immunofluorescence, using appropriate anti-tag antibodies. As shown in Fig. 1A, singly transfected cells gave positive immunofluorescence signals with anti-FLAG (Sig1R-FLAG) or anti-HA (Nav1.5-HA) antibodies, indicating the successful expression of the two proteins. In contrast, an anti-V5 antibody gave only a background signal (data not shown). In doubly transfected cells, the anti-HA and anti-FLAG signals extensively overlapped (Fig. 1B), indicating that the majority of transfected cells expressed both proteins. Again, an anti-V5 antibody gave only a background signal (data not shown). The reticular staining patterns suggest that both proteins were localized predominantly in the endoplasmic reticulum.
FIGURE 1.
Expression of the Sig1R and Nav1.5 in transfected tsA 201 cells. A, cells were singly transfected with either Sig1R-FLAG (left panel) or Nav1.5-HA (right panel). Cells were fixed, permeabilized and incubated with either mouse monoclonal anti-FLAG or mouse monoclonal anti-HA antibodies, followed by a Cy3-conjugated goat anti-mouse secondary antibody. Cells were imaged by confocal laser scanning microscopy. B, cells were co-transfected with Sig1R-FLAG and Nav1.5-HA. Cells were fixed, permeabilized, and incubated with both mouse monoclonal anti-FLAG and rabbit polyclonal anti-HA antibodies, followed by fluorescein isothiocyanate-conjugated goat anti-mouse and Cy3-conjugated goat anti-rabbit secondary antibodies. Cells were imaged by confocal laser scanning microscopy. Scale bar, 50 μm.
It is known that HEK-293 cells endogenously express the Sig1R (3). To assess the relative expression levels of endogenous and exogenous (FLAG-tagged) receptors in the tsA 201 cells, detergent extracts of Sig1R-FLAG-transfected and control cells were subjected to immunoblot analysis. The transfected cells gave strong signals with both an anti-FLAG antibody and an anti-Sig1R antibody (Fig. 2A). In contrast, the non-transfected cells gave no detectable signal with either antibody. Hence, endogenous Sig1R must be expressed at a much lower level than FLAG-tagged Sig1R and is therefore unlikely to interfere with the subsequent experiments.
FIGURE 2.
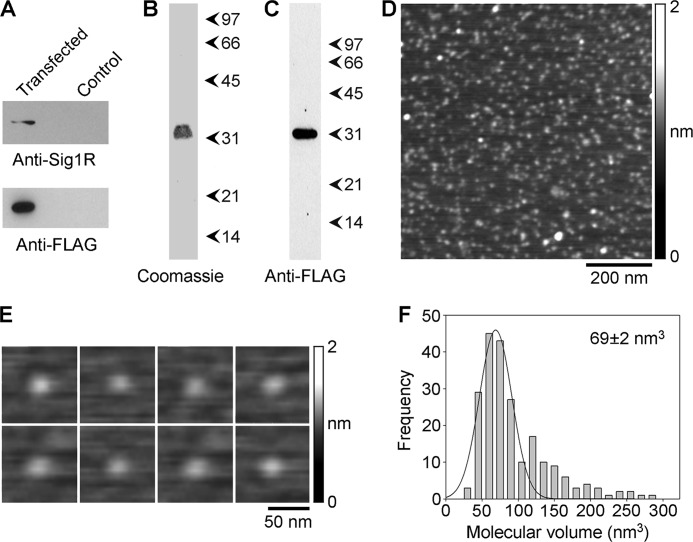
Isolation and AFM imaging of the Sig1R from singly transfected tsA 201 cells. A, detergent extracts of Sig1R-FLAG-transfected and control cells were analyzed by SDS-PAGE followed by immunoblotting using either a mouse monoclonal anti-FLAG antibody (bottom panel) or a rabbit polyclonal anti-Sig1R antibody (top panel). B and C, samples of protein isolated by immunoaffinity chromatography on anti-FLAG-agarose were analyzed by SDS-PAGE followed by either Coomassie Blue staining (B) or immunoblotting using a mouse monoclonal anti-FLAG antibody (C). Arrowheads indicate molecular mass markers (kDa). D, low-magnification AFM image of a sample of isolated Sig1R. E, gallery of enlarged images of Sig1R particles. F, frequency distribution of volumes of Sig1R particles. The curve indicates the fitted Gaussian function. The peak of the distribution is indicated.
Cells expressing Sig1R-FLAG alone were solubilized in Triton X-100 detergent (1% w/v), and the protein was isolated through the binding of the FLAG tag to anti-FLAG-agarose beads, followed by elution with a triple-FLAG peptide. A Coomassie Blue-stained gel of the isolated protein (Fig. 2B) shows a single major band at a molecular mass of 33 kDa. The isolated protein was also analyzed by immunoblotting using an anti-FLAG antibody (Fig. 2C). A single immunopositive band was seen, again at 33 kDa. This result demonstrates the successful isolation of Sig1R-FLAG from the transfected cells.
Low-magnification AFM images of isolated Sig1R-FLAG revealed a relatively homogenous distribution of particles (Fig. 2D). A gallery of zoomed images of individual particles is shown in Fig. 2E. The molecular volumes of a number of these particles were calculated, using Scanning Probe Image Processor. A frequency distribution of the volumes had a single peak, at 69 ± 2 (SE) nm3 (n = 216), close to the expected volume of 63 nm3 for a Sig1R, of a molecular mass of 33 kDa, according to Equation 2 (Fig. 2F). Hence, the imaged particles represent individual Sig1Rs.
Protein was next isolated from cells co-expressing Sig1R-FLAG and Nav1.5-HA through the binding of the HA tag on Nav1.5 to anti-HA beads, followed by elution with HA peptide. The isolated sample was analyzed by immunoblotting with anti-FLAG and anti-HA antibodies. The anti-FLAG antibody detected a single band at 33 kDa, as expected for the Sig1R (Fig. 3A, left panel), and the anti-HA blot revealed a single band at 260 kDa, as expected for Nav1.5 (Fig. 3A, right panel). The fact that the Sig1R was co-isolated with Nav1.5 indicates an intimate association between the two proteins.
FIGURE 3.
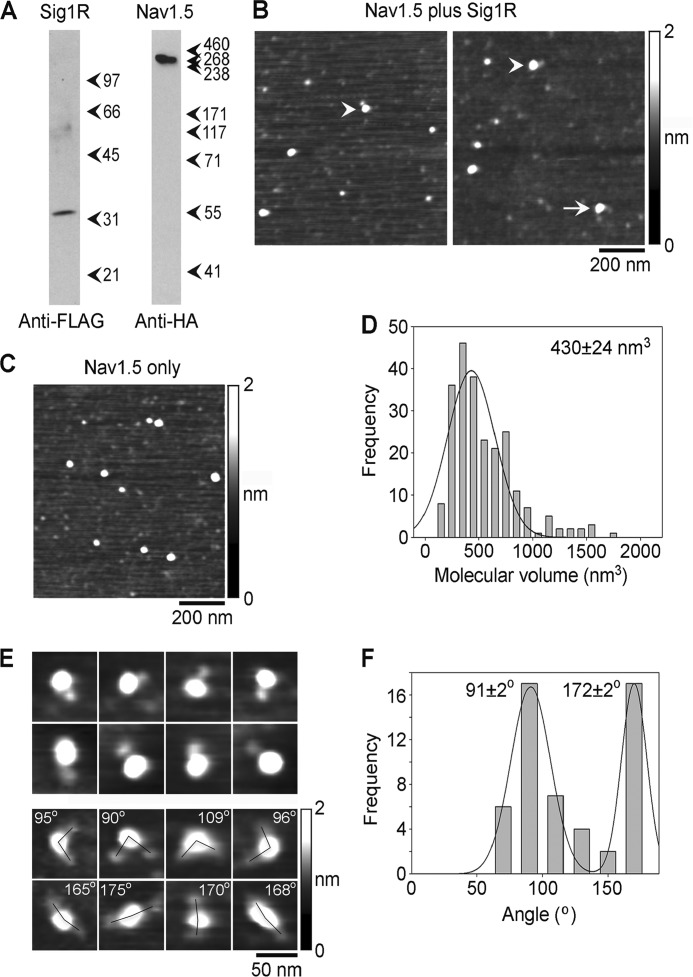
Isolation and analysis of Sig1R-FLAG·Nav1.5-HA complexes by immunoaffinity chromatography on anti-HA agarose. A, samples of protein isolated by immunoaffinity chromatography were analyzed by SDS-PAGE followed by immunoblotting using either mouse monoclonal anti-FLAG (left panel) or mouse monoclonal anti-HA antibodies (right panel). Arrowheads indicate molecular mass markers (kDa). B, low-magnification AFM images of samples of isolated Sig1R-FLAG/Nav1.5. Singly and doubly decorated large particles are indicated by arrowheads and an arrow, respectively. C, low-magnification AFM image of Nav1.5 alone, isolated by immunoaffinity chromatography on anti-HA-agarose. D, frequency distribution of volumes of large particles isolated from co-transfected cells that were decorated by Sig1R particles. The curve indicates the fitted Gaussian function. The peak of the distribution is indicated. E, gallery of zoomed images of Nav1.5 channels that were decorated by either one (upper panels) or two Sig1Rs (lower panels). Angles between pairs of bound Sig1Rs are indicated. F, frequency distribution of angles between pairs of bound Sig1Rs. The curve indicates the fitted Gaussian functions. The peaks of the distribution are indicated.
Low-magnification AFM images of co-isolated Sig1R and Nav1.5 showed a population of large particles, some of which were decorated by one (arrowheads) or two (arrow) smaller particles (Fig. 3B). In contrast, corresponding images of protein isolated from cells expressing Nav1.5 alone showed fewer decoration events (Fig. 3C). This result suggests that the small bound particles are Sig1Rs and the large particles are Nav1.5 channels.
To quantitate decoration of the Nav1.5 channels by the Sig1Rs, we set a volume range between 30 and 120 nm3, based on the volume of particles seen with the Sig1R alone (Fig. 2F). Of 2482 large particles, 143 (5.8%) were singly decorated by Sig1Rs, and of 6473 particles, 55 (0.8%) were doubly decorated. Corresponding percentages for Nav1.5 alone were as follows: 1.7% (56 of 3,244) singly decorated and 0.1% (7 of 5,917) doubly decorated. Hence, the vast majority of the decoration events seen were specific. A frequency distribution of volumes of the decorated central particles (Fig. 3D) had a single peak at 430 ± 24 nm3 (n = 231), close to the expected volume of 490 nm3 for a Nav1.5 particle with a molecular mass of 260 kDa. This result further supports the suggestion that these particles were indeed Sig1R-decorated Nav1.5 channels.
Galleries of zoomed images of singly and doubly decorated large particles are shown in Fig. 3E. We identified Nav1.5 channels that had been decorated by two Sig1Rs and measured the angles between the bound receptors. This was done in each case by joining the highest point on the central particle (the Nav1.5 channel) to the highest points on the peripheral particles (the Sig1Rs) by lines and then determining the angle between the two lines. A frequency distribution of the angles obtained is shown in Fig. 3F. The angle distribution has two peaks: a large peak at 91 ± 2° (n = 34) and a smaller peak at 172 ± 2° (n = 19); the ratio of the numbers of particles in the two peaks is 1.8:1. This angle profile, with two peaks at ∼90° and 180°, in a ratio of ∼2:1 suggests that the Nav1.5 presents four perpendicular binding sites to the Sig1R and that these are randomly occupied.
To further characterize the interaction between Sig1R and Nav1.5, we isolated protein from co-transfected cells through the binding of the FLAG tag on the Sig1R to anti-FLAG agarose, followed by elution with the triple-FLAG peptide. The isolated protein was analyzed by immunoblotting. As shown in Fig. 4A, the anti-FLAG blot showed a band at 33 kDa (left panel), and the anti-HA blot showed a band at 260 kDa (right panel). These results are very similar to those obtained after purification through the HA tag on Nav1.5 and support our assertion that the Sig1R and Nav1.5 interact intimately within the cells.
FIGURE 4.
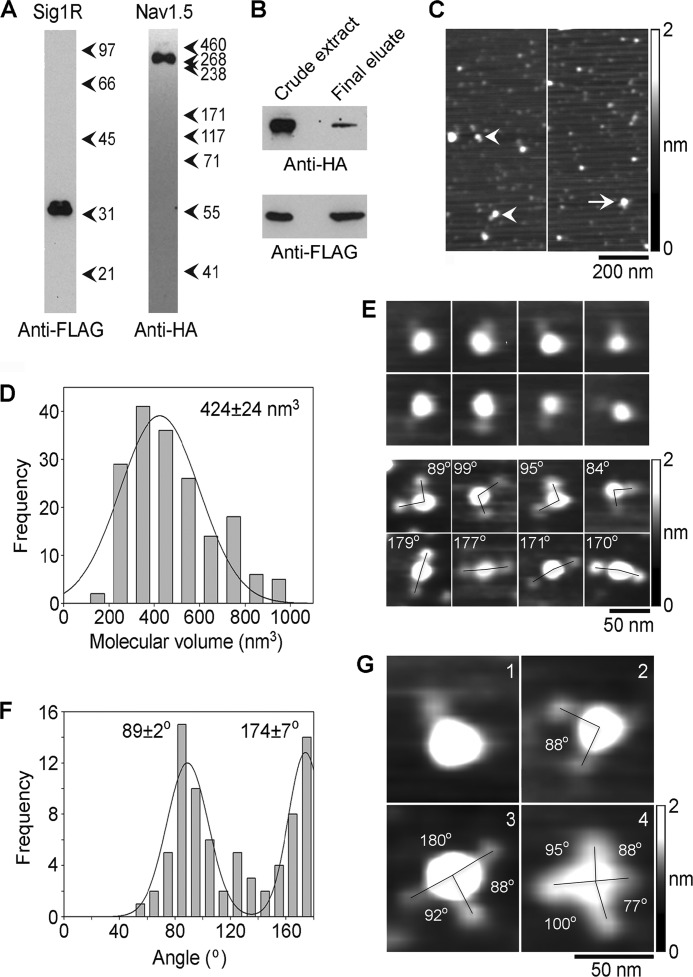
Isolation and analysis of Sig1R-FLAG·Nav1.5-HA complexes by immunoaffinity chromatography on anti-FLAG-agarose. A, samples of protein isolated by affinity chromatography were analyzed by SDS-PAGE followed by immunoblotting using either mouse monoclonal anti-FLAG (left panel) or mouse monoclonal anti-HA antibodies (right panel). Arrowheads indicate molecular mass markers (kDa). B, a sample of protein isolated from surface-biotinylated, intact co-transfected cells by sequential affinity chromatography on monomeric avidin-agarose and anti-FLAG-agarose was analyzed by SDS-PAGE followed by immunoblotting using either mouse monoclonal anti-FLAG (bottom panel) or mouse monoclonal anti-HA antibodies (top panel). A crude detergent extract of the cells (at a loading ratio of 1:128 compared with the final eluate) was also analyzed. C, low-magnification AFM images of samples of isolated Sig1R-FLAG/Nav1.5. Singly- and doubly-decorated large particles are indicated by arrowheads and an arrow, respectively. D, frequency distribution of volumes of large particles that were decorated by Sig1R particles. The curve indicates the fitted Gaussian function. The peak of the distribution is indicated. E, gallery of enlarged images of Nav1.5 channels that were decorated by either one (upper panels) or two Sig1Rs (lower panels). Angles between pairs of bound Sig1Rs are indicated. F, frequency distribution of angles between pairs of bound Sig1Rs. The curve indicates the fitted Gaussian functions. The peaks of the distribution are indicated. G, gallery of zoomed images of Nav1.5 particles decorated by one, two, three, or four Sig1R particles. Angles between pairs of bound Sig1Rs are indicated.
Given that the immunofluorescence images (Fig. 1) indicated that the majority of exogenously expressed protein was intracellular, whereas Nav1.5 clearly functions at the plasma membrane, we felt that it was important to establish whether the Sig1R and Nav1.5 interacted at the plasma membrane. To do this, we biotinylated proteins in the plasma membrane of intact co-transfected cells before preparing a detergent extract. Biotinylated proteins in the extract were captured on monomeric avidin-agarose and then eluted using free biotin. Biotinylated Sig1R-FLAG was then recaptured using anti-FLAG-agarose, followed by elution with triple-FLAG peptide. As shown in Fig. 4B, immunoblotting detected both Sig1R-FLAG and Nav1.5-HA in the final eluate, demonstrating an interaction between the two proteins when at least one of them was in the plasma membrane. To control for cell lysis during biotinylation, immunoblotting for β-actin was carried out; β-actin was detected in the total extract but not in the eluate from the avidin-agarose (data not shown), indicating that there was no significant cell lysis.
Low-magnification AFM images of protein isolated from a total cell extract showed a population of large particles some of which were decorated by either one (arrowheads) or two (arrow) smaller particles (Fig. 4C). In contrast, as shown above in Fig. 2D, an image of proteins isolated from cells singly transfected with the Sig1R alone shows very few larger particles.
As above, small bound particles in the volume range 30–120 nm3 were accepted as Sig1Rs. A frequency distribution of molecular volumes of the large decorated particles was then constructed. As shown in Fig. 4D, the distribution has a single peak at 424 ± 24 nm3 (n = 177), close to the value obtained for particles isolated through the HA tag on Nav1.5 (430 nm3, Fig. 3D) and to the value of 490 nm3 expected for a protein of molecular mass 260 kDa.
Galleries of zoomed images of singly- and doubly-decorated large particles are shown in Fig. 4E. A frequency distribution of angles between pairs of bound Sig1Rs (Fig. 4F) had two peaks, at 89 ± 2° (n = 49) and 174 ± 7° (n = 28), and the ratio of the sizes of the two peaks was 1.7:1, close to the values obtained in the reverse purification (91° and 172°, Fig. 3F) and to the predicted value of 2:1 for random decoration of a protein with 4-fold symmetry. Rarely, we saw Nav1.5 particles that were decorated by three (n = 4) or four (n = 2) Sig1R particles. Fig. 4G shows a gallery of zoomed images of Nav1.5 particles decorated by one, two, three, or four Sig1R particles. This gallery nicely illustrates the central conclusion of our study.
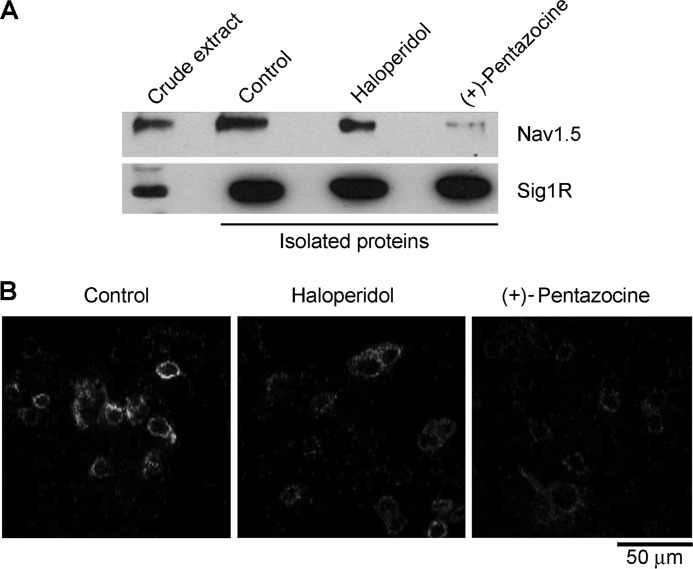
A number of Sig1R ligands are known to affect the function of ion channels, including Nav1.5 (3, 4, 23). We have shown previously that the ligand haloperidol reduced decoration of ASIC1a by the Sig1R by ∼50% (27). We therefore decided to test the effects of two Sig1R ligands, haloperidol and (+)-pentazocine, on the ability of Sig1R to capture Nav1.5 from a detergent extract of co-transfected cells. Drugs were incubated with the detergent extracts for 1 h at 4 °C, before the extracts were added to the immunobeads. As shown in Fig. 5A, neither ligand had any effect on the binding of the Sig1R to anti-FLAG agarose; however, both reduced the co-isolation of Nav1.5. The intensity of the Nav1.5 band was reduced to 80% of control by haloperidol and to 10% of control by (+)-pentazocine. Hence, both ligands reduce the interaction between the Sig1R and Nav1.5 when added in vitro.
FIGURE 5.
Effect of Sig1R ligands on the interaction between the Sig1R and Nav1.5. A, the Sig1R was isolated from Sig1R-FLAG/Nav1.5-HA co-transfected cells by immunoaffinity chromatography on anti-FLAG agarose. The Sig1R ligands haloperidol (Sigma; 100 μm) and (+)-pentazocine (Sigma; 10 μm) were incubated with identical samples of a crude detergent extract of the cells for 1 h at 4 °C before addition to the immunobeads. Samples of isolated protein were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting using either mouse monoclonal anti-HA (top panel) or anti-FLAG antibodies (bottom panel). The immunoblots shown are representative of the results from three separate experiments. A crude detergent extract of the cells (at a loading ratio of 1:32 compared with the final eluate) was also analyzed. B, cells were co-transfected with DNA encoding Nav1.5-HA and Sig1R-FLAG. Haloperidol (100 μm) and (+)-pentazocine (10 μm) were added to the media for 1 h before the cells were fixed. Cells were permeabilized and incubated with primary antibodies (rabbit polyclonal anti-HA and mouse monoclonal anti-FLAG) followed by anti-mouse (+) and anti-rabbit (−) proximity ligation secondary antibodies. The proximity ligation assay was then carried out and treated cells were imaged by confocal laser scanning microscopy. All images were captured using identical microscope settings. Scale bar, 50 μm.
To establish whether haloperidol and (+)-pentazocine also reduce the interaction between Nav1.5 and Sig1R within the tsA 201 cells, in situ proximity ligation assays were carried out (36). The assay uses two secondary antibodies, each bearing a short DNA strand. When the secondary antibodies are brought into close proximity (<40 nm) by binding to their relevant primary antibodies, the DNA strands hybridize with an additional circle-forming oligodeoxynucleotide. Ligation then creates a complete circularized oligodeoxynucleotide, and rolling circle amplification increases the amount of circular DNA several hundredfold. The DNA is then visualized using a fluorescent probe. Cells were co-transfected with Nav1.5-HA and Sig1R-FLAG, and drugs were added to the medium 1 h before the cells were fixed. Primary antibodies used to tag the subunits were rabbit anti-HA (Nav1.5) and mouse anti-FLAG (Sig1R). As shown in Fig. 5B, a strong proximity signal was seen in the control sample, indicating that Nav1.5 and Sig1R come into close proximity within the co-transfected cells. The intensity of the proximity signal was reduced by both haloperidol and (+)-pentazocine, with the latter drug having the greater effect. Hence, the two ligands have similar effects on the Nav1.5/Sig1R interaction in vitro and in intact cells.
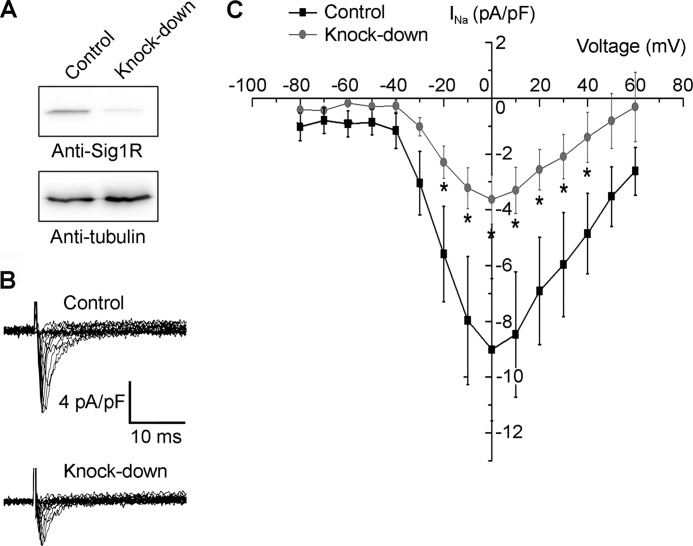
The experiments described thus far have used overexpressed, epitope-tagged Nav1.5 and Sig1R. We were therefore interested to examine whether Nav1.5 and Sig1R would also interact when expressed endogenously and whether such an interaction would have functional consequences for Nav1.5 channel behavior. To do this, we recorded Nav1.5 currents in control MDA-MB-231 cells and in cells in which Sig1R expression had been knocked down using shRNA; knockdown was confirmed by immunoblotting (Fig. 6A). The membrane potential was depolarized from a holding potential at −100 mV to potentials between −90 and +60 mV. A strong reduction of voltage-dependent Na+ current was observed in Sig1R knockdown cells compared with control cells (Fig. 6, B and C). Specifically, at 0 mV, the maximal current amplitude was −3.63 ± 0.84 pA/pF in Sig1R-silenced cells, and −9.01 ± 2.54 pA/pF in control cells (n = 10 cells for each condition). Thus, the knockdown of Sig1R expression in MDA-MB-231 cells reduces the Nav1.5 current by ∼60%, demonstrating that the Sig1R regulates Nav1.5 function in these cells.
FIGURE 6.
Sig1R expression regulates Nav1.5 current density in MDA-MB-231 cells. A, immunoblot showing the shRNA-induced knock-down of Sig1R expression in MDA-MB-231 cells. α-Tubulin is shown as a control (antibody from Sigma). B, representative Na+ currents elicited in Sig1R knockdown or control MDA-MB-231 cells by a series of depolarization pulses between +60 and −80 mV from a holding potential of −100 mV. C, current-voltage relationship of the Na+ current obtained from a holding potential of −100 mV (n = 10 cells). *, p < 0.05, Mann-Whitney test.
DISCUSSION
In the present study, we have shown that the Sig1R binds to Nav1.5, which contains four six-TMR cassettes, with 4-fold symmetry. In a previous study, we used a similar approach to demonstrate that the Sig1R binds to the trimeric ASIC1a ion channel with 3-fold symmetry (27). Of course, the fact that Nav1.5 contains all four TMR cassettes in a single polypeptide means that an intact channel has only one N- and one C- terminal domain. This would appear to rule out the possibility that either of these domains is involved in Sig1R binding, suggesting instead the likelihood that the Sig1R binds to the TMRs of its target proteins. This suggestion is supported by a recent study, which showed that the Sig1R could be co-immunoprecipitated with a truncation mutant of Kv1.3 that was essentially pared down to its TMRs (9).
If the Sig1R does indeed bind to the TMRs of its targets, the interaction is likely to be hydrophobic. There is no obvious structural motif shared by all Sig1R targets, and the Sig1R itself contains no known protein interaction motifs, such as SH3, PDZ, or WW domains (5). Significantly, two so-called sterol binding-like domains have been identified in the Sig1R (38, 39). One of these domains (residues 91–109) encompasses part of the second transmembrane domain, and the other (residues 176–194) forms a hydrophobic region close to the C terminus. The C-terminal hydrophobic region contains cholesterol binding domain motifs (VEYGR and LFYTLRSYAR), and as expected, this region binds cholesterol (40). Consistent with an involvement of cholesterol in the interaction between the Sig1R and its ion channel targets, both the Sig1R itself (18, 40) and ion channels such as Nav1.5 (31), Kv1.4 (41), and ASIC3 (42) are localized to cholesterol-enriched membrane regions, sometimes known as lipid “rafts.” The Sig1R ligand SKF-10047 has been shown to strongly inhibit cholesterol binding to the Sig1R and at the same time to reduce the raft localization of the receptor (40), suggesting that the cholesterol-binding domain forms part of the drug binding site. The observed effects of the two Sig1R ligands used here on the interaction between the Sig1R and Nav1.5 suggest that the action of the drugs might be mediated through displacement of cholesterol from the Sig1R. This possibility now needs to be tested in further studies.
Only a small proportion of the Nav1.5 particles isolated from cells co-expressing Nav1.5 and the Sig1R (∼6%) was decorated by Sig1Rs, and most decorated channels had only a single associated Sig1R. This result could indicate that the Sig1R interacts with only a subpopulation of the Nav1.5 channels within the cell, possibly at a particular stage of the intracellular transport pathway. Alternatively, the interaction between Nav1.5 and the Sig1R could be transient and/or sensitive to the addition of detergent used in the isolation protocol. The robust signal seen in the proximity ligation assay indicates that the two proteins do interact in the intact cell. Furthermore, the surface biotinylation experiment shows that the proteins interact when at least one of them is present in the plasma membrane. However, the stoichiometry of the interaction between the Sig1R and Nav1.5 in intact cells remains unresolved.
It has been reported previously that Nav1.5 promotes the invasiveness of the aggressive breast cancer cell line, MDA-MB-231 (29–31). Furthermore, the Sig1R was also found to be overexpressed in this cell line compared with a normal epithelial breast cell line (20, 40). Here, we have shown that Sig1R expression regulates Nav1.5 function in MDA-MB-231 cells. A similar observation was reported recently for the Sig1R regulation of the maturation and membrane stability of the heterotetrameric K+ channel, hERG, in leukemia cells (11), although further studies are needed to determine whether Sig1R regulates hERG and Nav1.5 through a common mechanism.
Voltage-gated Na+ channels such as Nav1.5 contain a pore-forming α-subunit and one or more β-subunits (28). Most of the experiments described here involved overexpression of the α-subunit in a cell line (tsA 201) that is unlikely to express a β-subunit. Consequently, we cannot completely rule out the possibility that the presence of a β-subunit might affect the interaction between the Sig1R and Nav1.5. However, we were able to observe a functional interaction between endogenous Sig1R and Nav1.5 in MDA-MB-231 cells, which are known to express Nav1.5 β-subunits (43). It is clear, therefore, that the Sig1R/Nav1.5 interaction does occur in the presence of the β-subunits.
In conclusion, we have demonstrated a direct interaction between the Sig1R and Nav1.5, revealed the architecture of the Sig1R·Nav1.5 complex, and shown that this interaction has functional consequences for Nav1.5 in a cancer cell line.
Acknowledgments
We thank Dr. Laurent Combettes (Université de Paris-Sud) for providing the MDA-MB-231 cells, Dr. C. Valdivia (University of Michigan) for providing the HA-tagged Nav1.5 construct, Dr. Patrick Martin (Université de Nice Sophia Antipolis) for generating the sh constructs, and Dr. M.B. Jackson (University of Wisconsin, Madison) for many constructive discussions.
This work was supported by the Jean Shanks Foundation and the James Baird Fund of the University of Cambridge (to A. P. S.), a Centre National de la Recherche Scientifique (CNRS) and Région Provence Alpes Côte d'Azur Fellowship (to D. C.), the University of Nice Sophia Antipolis, the CNRS, and the Association Ti'Toine Normandie (to O. S.), and the Wellcome Trust (to J. M. E.).
- Sig1R
- sigma-1 receptor
- AFM
- atomic force microscopy
- ASIC
- acid-sensing ion channel
- hERG
- human ether-à-go-go-related gene
- TMR
- transmembrane region.
REFERENCES
- 1. Monnet F. P. (2005) Sigma-1 receptor as regulator of neuronal intracellular Ca2+: clinical and therapeutic relevance. Biol. Cell 97, 873–883 [DOI] [PubMed] [Google Scholar]
- 2. Su T. P., Hayashi T., Maurice T., Buch S., Ruoho A. E. (2010) The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 31, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johannessen M., Ramachandran S., Riemer L., Ramos-Serrano A., Ruoho A. E., Jackson M. B. (2009) Voltage-gated sodium channel modulation by σ-receptor in cardiac myocytes and heterologous systems. Am. J. Physiol. Cell Physiol. 296, C1049-C1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johannessen M., Fontanilla D., Mavlyutov T., Ruoho A. E., Jackson M. B. (2011) Antagonist action of progesterone at σ-receptors in the modulation of voltage-gated sodium channels. Am. J. Physiol. Cell Physiol. 300, C328–C337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aydar E., Palmer C. P., Klyachko V. A., Jackson M. B. (2002) The sigma receptor as a ligand-regulated auxiliary potassium channel subunit. Neuron 34, 399–410 [DOI] [PubMed] [Google Scholar]
- 6. Kennedy C., Henderson G. (1990) Inhibition of potassium currents by the sigma receptor ligand (+)-3-(3-hydroxyphenyl)-N-(1-propyl)piperidine in sympathetic neurons of the mouse isolated hypogastric ganglion. Neuroscience 35, 725–733 [DOI] [PubMed] [Google Scholar]
- 7. Wilke R. A., Mehta R. P., Lupardus P. J., Chen Y., Ruoho A. E., Jackson M. B. (1999) σ Receptor photolabeling and σ receptor-mediated modulation of potassium channels in tumor cells. J. Biol. Chem. 274, 18387–18392 [DOI] [PubMed] [Google Scholar]
- 8. Zhang H., Cuevas J. (2005) σ Receptor activation blocks potassium channels and depresses neuroexcitability in rat intracardiac neurons. J. Pharmacol. Exp. Ther. 313, 1387–1396 [DOI] [PubMed] [Google Scholar]
- 9. Kinoshita M., Matsuoka Y., Suzuki T., Mirrielees J., Yang J. (2012) The sigma-1 receptor alters the kinetics of Kv1.3 voltage gated potassium channels but not the sensitivity to receptor ligands. Brain Res. 1452, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Cuevas J. (2002) Sigma receptors inhibit high-voltage-activated calcium channels in rat sympathetic and parasympathetic neurons. J. Neurophysiol. 87, 2867–2879 [DOI] [PubMed] [Google Scholar]
- 11. Crottès D., Martial S., Rapetti-Mauss R., Pisani D. F., Loriol C., Pellissier B., Martin P., Chevet E., Borgese F., Soriani O. (2011) Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. J. Biol. Chem. 286, 27947–27958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monnet F. P., Debonnel G., Junien J. L., De Montigny C. (1990) N-methyl-D-aspartate-induced neuronal activation is selectively modulated by sigma receptors. Eur. J. Pharmacol. 179, 441–445 [DOI] [PubMed] [Google Scholar]
- 13. Monnet F. P., Morin-Surun M. P., Leger J., Combettes L. (2003) Protein kinase C-dependent potentiation of intracellular calcium influx by sigma1 receptor agonists in rat hippocampal neurons. J. Pharmacol. Exp. Ther. 307, 705–712 [DOI] [PubMed] [Google Scholar]
- 14. Kume T., Nishikawa H., Taguchi R., Hashino A., Katsuki H., Kaneko S., Minami M., Satoh M., Akaike A. (2002) Antagonism of NMDA receptors by sigma receptor ligands attenuates chemical ischemia-induced neuronal death in vitro. Eur. J. Pharmacol. 455, 91–100 [DOI] [PubMed] [Google Scholar]
- 15. Hayashi T., Kagaya A., Takebayashi M., Shimizu M., Uchitomi Y., Motohashi N., Yamawaki S. (1995) Modulation by sigma ligands of intracellular free Ca2+ mobilization by N-methyl-d-aspartate in primary culture of rat frontal cortical neurons. J. Pharmacol. Exp. Ther. 275, 207–214 [PubMed] [Google Scholar]
- 16. Renaudo A., L'Hoste S., Guizouarn H., Borgèse F., Soriani O. (2007) Cancer cell cycle modulated by a functional coupling between sigma-1 receptors and Cl− channels. J. Biol. Chem. 282, 2259–2267 [DOI] [PubMed] [Google Scholar]
- 17. Herrera Y., Katnik C., Rodriguez J. D., Hall A. A., Willing A., Pennypacker K. R., Cuevas J. (2008) σ-1 receptor modulation of acid sensing ion channel a (ASIC1a) and ASIC1a-induced Ca2+ influx in rat cortical neurons. J. Pharmacol. Exp. Ther. 327, 491–502 [DOI] [PubMed] [Google Scholar]
- 18. Hayashi T., Su T. P. (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 19. Lysko P. G., Gagnon R. C., Yue T. L., Gu J. L., Feuerstein G. (1992) Neuroprotective effects of SKF 10,047 in cultured rat cerebellar neurons and in gerbil global brain ischemia. Stroke 23, 414–419 [DOI] [PubMed] [Google Scholar]
- 20. Aydar E., Onganer P., Perrett R., Djamgoz M. B., Palmer C. P. (2006) The expression and functional characterization of sigma 1 receptors in breast cancer cell lines. Cancer Lett. 242, 245–257 [DOI] [PubMed] [Google Scholar]
- 21. Spruce B. A., Campbell L. A., McTavish N., Cooper M. A., Appleyard M. V., O'Neill M., Howie J., Samson J., Watt S., Murray K., McLean D., Leslie N. R., Safrany S. T., Ferguson M. J., Peters J. A., Prescott A. R., Box G., Hayes A., Nutley B., Raynaud F., Downes C. P., Lambert J. J., Thompson A. M., Eccles S. (2004) Small molecule antagonists of the sigma-1 receptor cause selective release of the death program in tumor and self-reliant cells and inhibit tumor growth in vitro and in vivo. Cancer Res. 64, 4875–4886 [DOI] [PubMed] [Google Scholar]
- 22. Hanner M., Moebius F. F., Flandorfer A., Knaus H. G., Striessnig J., Kempner E., Glossmann H. (1996) Purification, molecular cloning, and the expression of the mammalian sigma-1 binding site. Proc. Natl. Acad. Sci. U.S.A. 93, 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontanilla D., Johannessen M., Hajipour A. R., Cozzi N. V., Jackson M. B., Ruoho A. E. (2009) The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 323, 934–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lupardus P. J., Wilke R. A., Aydar E., Palmer C. P., Chen Y., Ruoho A. E., Jackson M. B. (2000) Membrane-delimited coupling between sigma receptors and K+ channels in rat neurohypophysial terminals requires neither G-proteins nor ATP. J. Physiol. 526, 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 26. Carnally S. M., Dev H. S., Stewart A. P., Barrera N. P., Van Bemmelen M. X., Schild L., Henderson R. M., Edwardson J. M. (2008) Direct visualization of the trimeric structure of the ASIC1a channel, using AFM imaging. Biochem. Biophys. Res. Commun. 372, 752–755 [DOI] [PubMed] [Google Scholar]
- 27. Carnally S. M., Johannessen M., Henderson R. M., Jackson M. B., Edwardson J. M. (2010) Demonstration of a direct interaction between sigma-1 receptors and acid-sensing ion channels. Biophys. J. 98, 1182–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catterall W. A., Goldin A. L., Waxman S. G. (2005) International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57, 397–409 [DOI] [PubMed] [Google Scholar]
- 29. Roger S., Besson P., Le Guennec J. Y. (2003) Involvement of a novel fast inward sodium current in the invasion capacity of a breast cancer cell line. Biochim. Biophys. Acta 1616, 107–111 [DOI] [PubMed] [Google Scholar]
- 30. Gillet L., Roger S., Besson P., Lecaille F., Gore J., Bougnoux P., Lalmanach G., Le Guennec J. Y. (2009) Voltage-gated sodium channel activity promotes cysteine cathepsin-dependent invasiveness and colony growth of human cancer cells. J. Biol. Chem. 284, 8680–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brisson L., Gillet L., Calaghan S., Besson P., Le Guennec J. Y., Roger S., Gore J. (2011) Nav1.5 enhances breast cancer cell invasiveness by increasing NHE1-dependent H+ efflux in caveolae. Oncogene 30, 2070–2076 [DOI] [PubMed] [Google Scholar]
- 32. Schneider S. W., Lärmer J., Henderson R. M., Oberleithner H. (1998) Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflügers Arch. 435, 362–367 [DOI] [PubMed] [Google Scholar]
- 33. Neaves K. J., Cooper L. P., White J. H., Carnally S. M., Dryden D. T., Edwardson J. M., Henderson R. M. (2009) Atomic force microscopy of the EcoKI type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res. 37, 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott D. W. (1979) On optimal and data-based histograms. Biometrika 66, 605–610 [Google Scholar]
- 35. Welch B. L. (1947) The generalization of student's problems when several different population variances are involved. Biometrika 34, 28–35 [DOI] [PubMed] [Google Scholar]
- 36. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 37. Albagli-Curiel O., Lécluse Y., Pognonec P., Boulukos K. E., Martin P. (2007) A new generation of pPRIG-based retroviral vectors. BMC Biotechnol. 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pal A., Hajipour A. R., Fontanilla D., Ramachandran S., Chu U. B., Mavlyutov T., Ruoho A. E. (2007) Identification of regions of the sigma-1 receptor ligand binding site using a novel photoprobe. Mol. Pharmacol. 72, 921–933 [DOI] [PubMed] [Google Scholar]
- 39. Pal A., Chu U. B., Ramachandran S., Grawoig D., Guo L. W., Hajipour A. R., Ruoho A. E. (2008) Juxtaposition of the steroid binding domain-like I and II regions constitutes a ligand binding site in the sigma-1 receptor. J. Biol. Chem. 283, 19646–19656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palmer C. P., Mahen R., Schnell E., Djamgoz M. B., Aydar E. (2007) Sigma-1 receptors bind cholesterol and remodel lipid rafts in breast cancer cell lines. Cancer Res. 67, 11166–11175 [DOI] [PubMed] [Google Scholar]
- 41. Wong W., Schlichter L. C. (2004) Differential recruitment of Kv1.4 and Kv4.2 to lipid rafts by PSD-95. J. Biol. Chem. 279, 444–452 [DOI] [PubMed] [Google Scholar]
- 42. Eshcol J. O., Harding A. M., Hattori T., Costa V., Welsh M. J., Benson C. J. (2008) Acid-sensing ion channel 3 (ASIC3) cell surface expression is modulated by PSD-95 within lipid rafts. Am. J. Physiol. Cell Physiol. 295, C732–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chioni A. M., Brackenbury W. J., Calhoun J. D., Isom L. L., Djamgoz M. B. (2009) A novel adhesion molecule in human breast cancer cells: voltage-gated Na+ channel β1 subunit. Int. J. Biochem. Cell Biol. 41, 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]