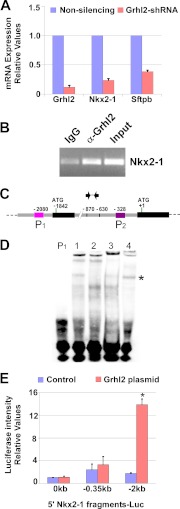
FIGURE 5.
GRHL2 binds to Nkx2-1 promoter and activates its transcription. A, qRT-PCR expression analysis of Nkx2-1 and its target Sftpb in MLE15 cells infected with Grhl2-shRNA or non-silencing-shRNA. Data sets were normalized against Gapdh (n = 3). Data represent the mean ± S.D. B, ChIP-PCR analysis shows GRHL2 binding to the Nkx2-1 promoter in the embryonic lung. ChIP was performed with chromatin from E11.5 day lung buds immunoprecipitated with GRHL2-specific antibody or its isotype control. Immunoprecipitated and input DNA fragments were amplified using primers corresponding to Nkx2-1. C, a scheme of the Nkx2-1 gene depicting 5′ regulatory regions, first and second exons (black boxes), and first intron (gray boxes). Numbers are relative to the second ATG (+1). Pink (P1) and purple boxes (P2) represent probes used in EMSAs. Arrows indicate oligonucleotides used in ChIP-PCR analyses. D, EMSA analysis shows binding of MLE15 nuclear proteins to probe P1 (lane 1), competition with 100-fold unlabeled probe (lane 2), interference of complex formation using GRHL2 antibody (lane 3), and control with IgG. The asterisk marks the P1-protein complex. E, the Nkx2-1 promoter is activated in the presence of exogenous GRHL2. MLE15 cells were transfected with either empty luciferase plasmid (pGL3; 0-Luc-plasmid), Nkx2-1 promoter luciferase plasmids −0.35kbNkx2-1Luc or −2.1KbNkx2-1Luc, and co-transfected with CMV-Grhl2-UBC-Gfp or CMV-dsred-UBC-Gfp plasmids. Firefly luciferase was normalized to renilla luciferase levels (n = 2–3). Data represent the mean ± S.D. The asterisk indicates p ≤ 0.05.