Background: Chronic hepatitis C virus (HCV) infection increases the risk of type 2 diabetes and hepatic steatosis.
Results: Phosphoenolpyruvate carboxykinase (PEPCK) and associated transcription factors are up-regulated in HCV-infected Huh.8 cells.
Conclusion: Increased CCAAT/enhancer-binding protein β (C/EBPβ) and nonstructural component 5A (NS5A) are essential components for increased gluconeogenesis.
Significance: NS5A and C/EBPβ may possibly be considered as a new pharmacological target during HCV infection.
Keywords: Diabetes, Gluconeogenesis, Insulin, Liver Metabolism, Obesity, NASH
Abstract
Chronic hepatitis C virus (HCV) infection greatly increases the risk for type 2 diabetes and nonalcoholic steatohepatitis; however, the pathogenic mechanisms remain incompletely understood. Here we report gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) transcription and associated transcription factors are dramatically up-regulated in Huh.8 cells, which stably express an HCV subgenome replicon. HCV increased activation of cAMP response element-binding protein (CREB), CCAAT/enhancer-binding protein (C/EBPβ), forkhead box protein O1 (FOXO1), and peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and involved activation of the cAMP response element in the PEPCK promoter. Infection with dominant-negative CREB or C/EBPβ-shRNA significantly reduced or normalized PEPCK expression, with no change in PGC-1α or FOXO1 levels. Notably, expression of HCV nonstructural component NS5A in Huh7 or primary hepatocytes stimulated PEPCK gene expression and glucose output in HepG2 cells, whereas a deletion in NS5A reduced PEPCK expression and lowered cellular lipids but was without effect on insulin resistance, as demonstrated by the inability of insulin to stimulate mobilization of a pool of insulin-responsive vesicles to the plasma membrane. HCV-replicating cells demonstrated increases in cellular lipids with insulin resistance at the level of the insulin receptor, increased insulin receptor substrate 1 (Ser-312), and decreased Akt (Ser-473) activation in response to insulin. C/EBPβ-RNAi normalized lipogenic genes sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor γ, and liver X receptor α but was unable to reduce accumulation of triglycerides in Huh.8 cells or reverse the increase in ApoB expression, suggesting a role for increased lipid retention in steatotic hepatocytes. Collectively, these data reveal an important role of NS5A, C/EBPβ, and pCREB in promoting HCV-induced gluconeogenic gene expression and suggest that increased C/EBPβ and NS5A may be essential components leading to increased gluconeogenesis associated with HCV infection.
Introduction
Hepatitis C virus (HCV)6 infection is one of the leading causes of liver disease, with an estimated 170 million infected individuals worldwide (1). HCV typically results in a prolonged, clinical course that may progress over decades toward liver fibrosis, cirrhosis, and eventually hepatocellular carcinoma (2, 3). HCV infection is strongly associated with nonalcoholic fatty liver disease and the development of diabetes (4–6). Mounting evidence suggests that HCV-associated insulin resistance may be an underlying cause of hepatic steatosis and progression to diabetes (7). Steatosis and impaired insulin receptor substrate 1 (IRS-1)/PI3K response have been observed in the liver of patients infected with HCV (8). However, the underlying mechanisms for HCV-induced diabetes remain to be fully defined.
Initially, the HCV core was implicated in the pathogenesis of steatosis (9, 10) and insulin resistance in patients with HCV infection (11); however, recent studies suggest that nonstructural component 5A (NS5A) represents a candidate protein that could contribute to the pathogenesis of lipid formation (12) and by extension insulin resistance. Whereas a role for NS5A in HCV-induced steatosis has been suggested, its role in the regulation of genes involved in hepatic glucose metabolism, particularly the rate-limiting gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK), remains unclear. Among the complex network of transcription factors and cofactors that contribute to gluconeogenic gene expression, forkhead box O1 (FOXO1), cAMP response element-binding protein (CREB), CCAAT/enhancer-binding proteins (C/EBP) α and β, and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) are particularly important effectors of the cAMP pathway. The prevailing model is that induction of gluconeogenic genes by glucagon or cAMP is mediated by phosphorylation of CREB, which must interact with C/EBPβ and other factors bound to an upstream accessory enhancer to stimulate gene transcription. Furthermore, CREB induces PGC-1α and via its interaction with hepatocyte nuclear factor 4α, plays a crucial role in PEPCK and glucose-6-phosphatase (G-6-Pase) transcriptional regulation (13). Insulin inhibits gluconeogenesis in part through nuclear exclusion of the phosphorylated form of FOXO1 (14, 15) and by decreasing its interaction with PGC-1α (16).
In addition to gluconeogenesis, previous data demonstrate that C/EBPβ may play a critical role in steatosis, lipid synthesis, inflammation, and possibly ER stress in mice (17–19). The contribution of C/EBPβ to activation of the gluconeogenic and lipogenic program in HCV cells has thus far not been addressed experimentally. In the present study, we provide novel information that HCV-replicating cells, in the absence of inflammatory cytokines, dramatically activate PEPCK gene expression and genes coding for lipid uptake and the de novo pathway for lipogenesis. This is accompanied by inhibition of insulin signaling and increased lipid accumulation, all important characteristics underlying the progression to nonalcoholic fatty liver disease. Our results reveal that both NS5A and C/EBPβ knockdown separately suppress several key genes important for gluconeogenesis and de novo lipogenesis, indicating that C/EBPβ, in addition to NS5A, may control genes critical for the progression to diabetes in HCV-infected cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
Rice and co-workers (20) developed an elegant in vitro cell culture-based system using subgenomic replicons of HCV. The HCV subgenomic replicon in Huh.8 cells is replication-competent, because it is able to synthesize minus-strand HCV RNA that serves as substrate for copying more plus strand genomic RNA (see Fig. 1). The generation and maintenance of wild type and stably infected Huh7 cells with HCV subgenomic replicon (Huh.8) has been described (20). Ava.1 cells contain a 47-amino acid deletion in the NS5A gene within the zinc-binding domain, designed to limit transcriptional activation (21). Huh7 cells were cultured in complete DMEM (4.5 g/L glucose) supplemented with 10% FBS. Huh.8 and Ava.1 cells were maintained in complete DMEM supplemented with 10% heat-inactivated FBS, nonessential amino acids, and 1 mg/ml G418. Primary hepatocytes were prepared using standardized methods described previously (22). All of the cells were maintained at 37 °C in 5% CO2.
FIGURE 1.
Pictorial view of HCV subgenomic replicon expressed in Huh.8 cells. Huh.8 cells contain the stable integration of HCV nonstructural components NS2, NS3, NS4A, NS4B, NS5A, and NS5B, whereas Ava.1 contains NS2, NS3, NS4A, NS4B, NS5B, and mutated NS5A with a 47-amino acid deletion.
Recombinant Adenoviruses and Plasmids
The NS5A adenovirus (Ad-NS5A) (23) has been described previously. The dominant-negative CREB adenovirus (Ad-ACREB) was constructed using ACREB cDNA provided by Dr. Charles Vinson (National Cancer Institute). The construction of −490-bp PEPCK promoter-LUC (PEPCK-LUC) and glucocorticoid response element (GRE) mutant PEPCK-LUC construct (−430-LUC mutant) have been described previously (24, 25). Oligonucleotide 5′-AGGCCGGCCTTAGTTACCCGAGGCGAGC-3′ was used to mutate the cAMP response element (CRE) site in PEPCK-LUC to create construct CRE mutant. The control plasmid pGL3-LUC was from Promega (Madison, WI). Luciferase activity was quantitated as previously described (23). C/EBPβ and nontargeting shRNA adenoviruses have been described (18).
Western Blot Analysis
Huh7 and Huh.8 cells were grown to 70% confluence. The cells were serum-starved for 3 h in DMEM and subsequently stimulated with 100 nm insulin for 10 min. The cells were washed with PBS and pelleted at 200 × g for 5 min. Cell pellets were resuspended in lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 2 mm EDTA, 2.5 mm sodium pyrophosphate, 20 mm β-glycerophosphate, 10% glycerol plus protease and phosphatase inhibitors), rocked at 4 °C for 30 min, and spun at 16,000 × g for 5 min to pellet insoluble material. Nuclear proteins from liver cells were prepared as described (23). Immunoblot assays were performed using 50 μg of total or nuclear protein or cytoplasmic extract as previously described (17) for the following antibodies: phospho-Akt (Ser-473), phospho-CREB (Ser-133), insulin receptor β (IR-β, Tyr-1146), Akt, tubulin, phospho-FOXO1 (Ser-256), FOXO1, and Bcl-2 (Cell Signaling Technology, Danvers, MA); IR-β, IRS-1, CREB, and C/EBPβ (Santa Cruz Biotechnology, Santa Cruz, CA); NS5A and TATA binding protein (Abcam, Cambridge, MA); and phospho-IRS-1 (Ser-307) for detection of IRS-1 (Ser-312) in humans from Millipore (Billerica, MA).
RNA Isolation and Quantitative Real Time PCR
Total RNA extraction, cDNA synthesis, and quantitative real time PCR (qPCR) were performed as previously described (26). RNA expression data were normalized to levels of reference gene ribosomal protein L13A or ubiquitin C using the comparative threshold cycle method.
Nile Red Staining
Huh7, Huh.8, and Ava.1 cells were seeded at a density of 5 × 104 cells/well. The cells were collected by trypsinization, washed in PBS, pooled with free floating cells collected from the culture medium, and fixed in 4% formaldehyde, freshly prepared from paraformaldehyde, in 0.1 m potassium phosphate, pH 7.2. The cells were stained with 1 mg/ml of Nile Red and viewed on a Nikon Optiphot-II epifluorescence microscope equipped with a Cool Snap Pro camera and quantified with Image Pro Plus software.
Triglyceride Analysis
Huh7 and Huh.8 cells were infected with 50 pfu/cell shRNA adenoviruses for 48 h. The lipids were extracted by butanol lipid extraction as described previously (27). Triglycerides (TGs) were quantified by detecting free glycerol after lipase treatment using TG reagent per manufacturer's directions (Sigma-Aldrich).
Glucose Output Assay
HepG2 cells were infected with Ad-GFP or Ad-NS5A for 48 h. The medium was replaced with serum-free medium overnight, and then glucose- and phenol red-free medium supplemented with 2 mm sodium pyruvate and 20 mm sodium lactate was added to the cells. The cells were 80–100% confluent. Medium samples were collected at 2, 4, and 6 h. After 6 h, the cells were washed with PBS and lysed for protein estimation. Glucose production was measured in medium samples with a glucose assay kit (Sigma).
Measurement of Exocytosis, Imaging, and Analysis
The rate of exocytosis was assessed by real time imaging using fluorescent probe FM1-43 (Invitrogen) as described (28). Upon the addition of FM1-43 to the external solution, the probe partitions into the plasma membrane exposed to the external solution. When vesicles fuse with the plasma membrane, FM1-43 equilibrates with the new membrane, resulting in an increase in the apparent fluorescence. Therefore, the overall change in cellular FM1-43 fluorescence provides a measure of the sum of all exocytotic events.
Statistical Analysis
The data are expressed as the means ± S.E. and were analyzed statistically by using one-way analysis of variance or Student's t test. A p value of < 0.05 was determined to be statistically significant.
RESULTS
Outline of the HCV Genome with Structural and Nonstructural Components
The Huh.8 cell line contains the HCV subgenomic replicon expressing nonstructural components (NS2, NS3, NS4A, NS4B, NS5A, and NS5B), whereas the Huh7/HCVrep1bBB1 Ava.1 cells contain a 47-amino acid deletion in the NS5A gene within the zinc-binding domain, designed to limit transcriptional activation (Fig. 1) (21).
HCV Protein Expression Increases PEPCK Gene Transcription and Glucose Production
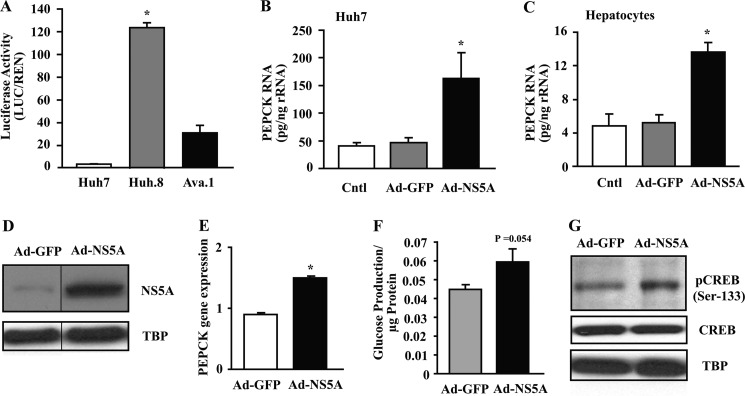
Because HCV infection alters glucose metabolism, we investigated the effect of HCV expression on PEPCK, a key regulator of gluconeogenesis. The majority of the control units necessary for tissue-specific and hormonal regulation of the hepatic PEPCK promoter are contained within the first −490 bp of the promoter (29). To observe the effect of HCV on PEPCK gene expression, we transfected the −490 bp PEPCK promoter-LUC construct (PEPCK-LUC) into Huh7, Huh.8, and Ava.1 cells, and luciferase activity was measured (Fig. 2A). The luciferase activity corrected for transfection efficiency was substantially higher in HCV-replicating Huh.8 cells versus nonreplicating Ava.1 cells with a mutant form of NS5A (∼20-fold). Using just the Ad-NS5A protein, we infected Huh7 cells and isolated primary rat hepatocytes. The results showed a robust ∼3-fold increase in endogenous PEPCK mRNA by qPCR, whereas no changes were seen with Ad-GFP (Fig. 2, B and C). These results demonstrate that although NS5A alone can induce PEPCK expression, mutating NS5A substantially reduces, but does not normalize, PEPCK gene expression in the context of HCV-replicating cells, suggesting that other viral components participate in the full activation process. To evaluate whether increased PEPCK was correlated with gluconeogenesis, HepG2 cells were infected with Ad-NS5A (Fig. 2D), and cellular glucose production was measured. First, PEPCK gene expression was analyzed, and NS5A-infected cells showed a 1.5-fold increase in endogenous PEPCK mRNA compared with Ad-GFP-infected cells (Fig. 2E). Importantly, Ad-NS5A cells demonstrated higher glucose production (p = 0.054; Fig. 2F). This was also associated with an increase in pCREB (Ser-133), with no change in CREB levels (Fig. 2G).
FIGURE 2.
HCV protein expression increases PEPCK gene transcription in a CRE-dependent manner. A, luciferase activity of PEPCK-LUC was measured in Huh7, Huh.8, and Ava.1 cells as outlined under “Experimental Procedures.” The luciferase activity was corrected for transfection efficiency. The values are presented as the means ± S.E. *, p < 0.05 (n = 3). B and C, PEPCK RNA levels were measured by qPCR in uninfected control cells and cells infected with either Ad-GFP or Ad-NS5A in both Huh7 cells and isolated primary rat hepatocytes, respectively. The values are presented as the means ± S.E. *, p < 0.05 (n = 3). D, equal amounts of nuclear extracts were collected from HepG2 cells transfected with either Ad-GFP or Ad-NS5A, and subjected to Western blot analysis with NS5A and TATA binding protein (TBP). Representative blots are presented. E, PEPCK mRNA levels were measured by qPCR in cells infected with either Ad-GFP or Ad-NS5A, and the data were normalized to ubiquitin C. *, p < 0.001 (n = 4). F, glucose production was measured in 2-h medium samples of HepG2 cells infected with Ad-GFP or Ad-NS5A (n = 4). The values are presented as the means ± S.E. p = 0.054. G, overexpression of NS5A increased CREB phosphorylation in HepG2 cell line. Equal amounts of nuclear extracts were collected from HepG2 cells infected with Ad-GFP and Ad-NS5A and subjected to Western blot analysis. Representative blots are presented.
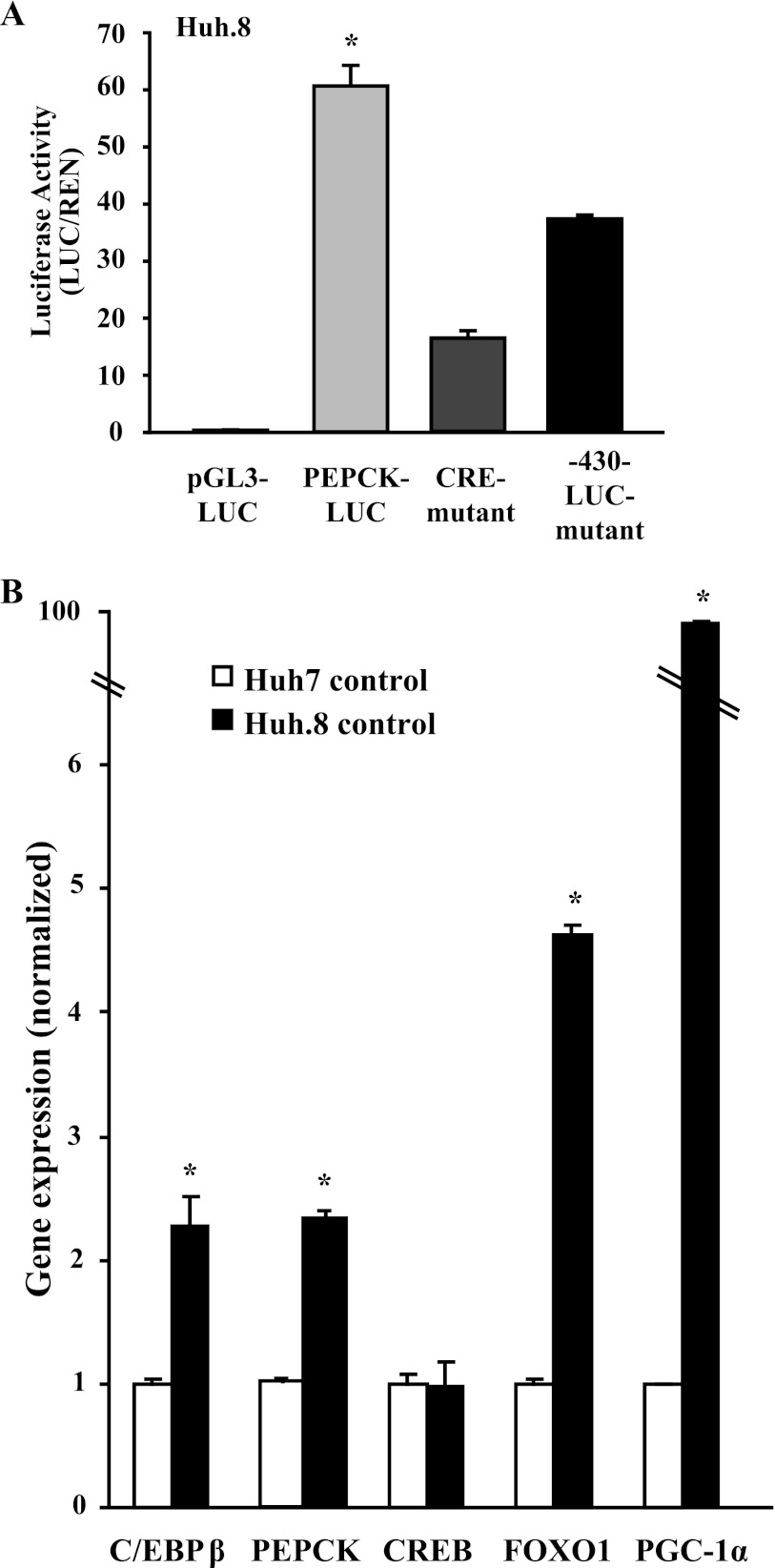
The higher PEPCK mRNA levels and increased pCREB in HCV-infected cells prompted us to examine the PEPCK promoter further. It is well established that both the GRE and the CRE are required for full cAMP-mediated expression of the PEPCK gene (13, 29). Mutant PEPCK-LUC constructs with a mutation in the CRE site (CRE mutant) or deletion of the GRE site (−430-LUC mutant) were assayed by transient transfection and compared with full-length PEPCK-LUC in HCV-expressing Huh.8 cells (Fig. 3A). PEPCK transcriptional activity was reduced 75% with the CRE mutant construct, suggesting that factors acting through this site had a major effect on induction of the PEPCK promoter. Deletion of the GRE site also reduced PEPCK promoter activity by 30%, suggesting that cooperation might exist between the GRE and CRE in the promoter, contributing to the increased PEPCK gene transcription in HCV-expressing cells.
FIGURE 3.
PEPCK promoter analysis and gene expression in Huh.8 cells. A, PEPCK transcriptional activity was measured in Huh.8 cells by DNA transfections of promoter-less control construct (pGL3-LUC), PEPCK-LUC, CRE mutant PEPCK-LUC (CRE mutant), and GRE mutant PEPCK-LUC (−430-LUC mutant). The values are presented as the means ± S.E. *, p < 0.05 (n = 3). B, comparison of gluconeogenic gene expression in Huh7 and Huh.8 cells. Transcription factor and coactivator gene expression were analyzed by qPCR in Huh7 and Huh.8 cells, and the data were normalized to reference gene RPL13A (n = 3–4). The values are presented as the means ± S.E. *, p < 0.05.
Transcription Factor Expression in Huh7 and Huh.8 Cells
Among the complex network of transcription factors and cofactors that regulate PEPCK gene expression, PGC-1α, FOXO1, CREB, and C/EBPβ are particularly important (30). PGC-1α does not directly bind to the PEPCK promoter but facilitates transcriptional activity of many factors (31). This background prompted us to examine their expression pattern in parental Huh7 cells compared with Huh.8 cells. C/EBPβ, FOXO1, and PGC-1α mRNA were significantly greater (∼2.5-, 5-, and > 10-fold increases, respectively) in Huh.8 cells compared with Huh7 (Fig. 3B). The endogenous PEPCK mRNA expression was ∼2.5-fold higher in nonstimulated Huh.8 cells as expected. Notably, HCV expression did not affect CREB mRNA levels.
Molecular Effects of Insulin on PEPCK and pFOXO1 Expression in NS5A-infected HepG2 Cells
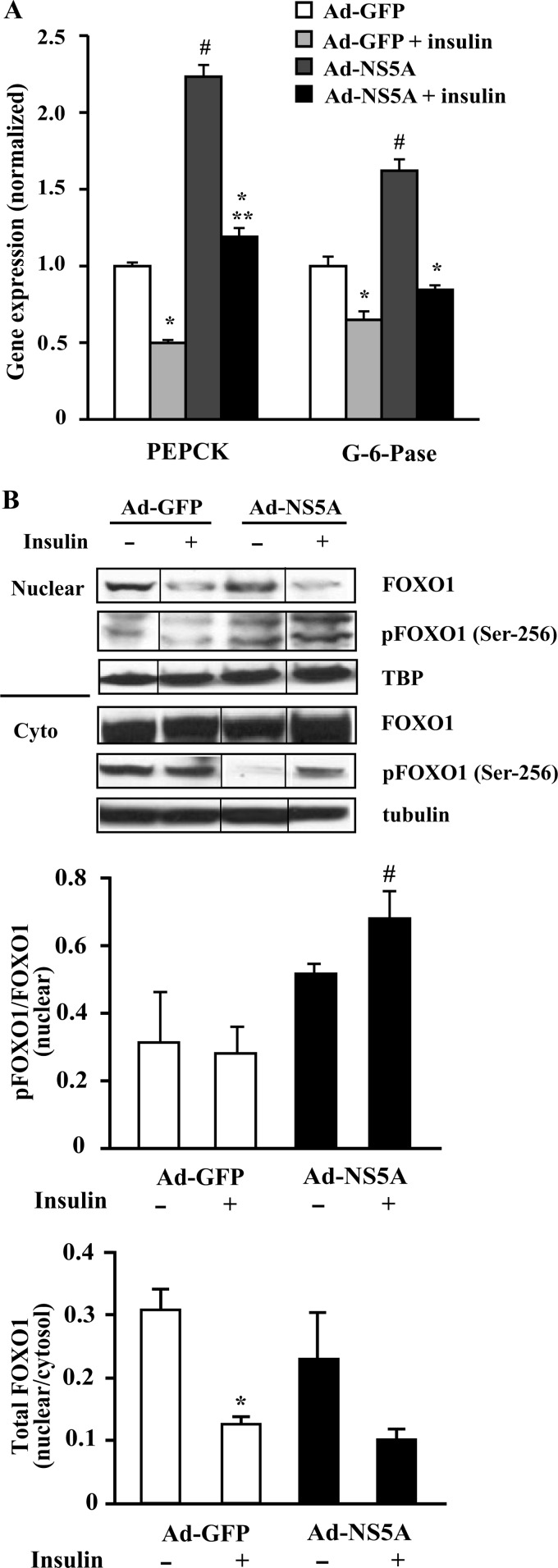
Insulin inhibition of gluconeogenic gene expression is the dominant mechanism for suppressing glucose production; however, the mechanisms of the effects of insulin on hepatic glucose metabolism with HCV infection are only recently becoming delineated. The balance between insulin suppression of glucose production and viral-induced activation of PEPCK is in favor of increased glucose production (32). As shown in Fig. 4A, insulin suppressed both PEPCK and G-6-Pase significantly in Ad-GFP-treated cells. In NS5A-infected cells, PEPCK and G-6-Pase expression were significantly increased compared with Ad-GFP-treated cells, and insulin significantly suppressed PEPCK and G-6-Pase mRNA relative to Ad-NS5A-treated cells. However, PEPCK expression in Ad-NS5A-treated cells remained significantly increased relative to insulin-treated Ad-GFP cells, whereas G-6-Pase expression trended toward a similar increase (p = 0.09).
FIGURE 4.
Insulin action on PEPCK, G-6-Pase, and pFOXO1 expression in NS5A-overexpressed HepG2 cells. HepG2 cells were infected with either Ad-GFP or Ad-NS5A and treated with or without 100 nm insulin for 6 h. A, PEPCK and G-6-Pase RNA levels were measured by qPCR. The values are presented as the means ± S.E. *, p < 0.05 versus no insulin; #, p < 0.005 versus Ad-GFP; **, p < 0.02 versus Ad-GFP + insulin (n = 3). B, equal amounts of nuclear/cytoplasmic (Cyto) extracts were collected from the similar treatment and subjected to Western blot analysis with pFOXO1 (Ser-256), total FOXO1, and TATA binding protein (TBP) or tubulin as a loading control. The representative blot is a composite of a larger blot in which all the samples were run simultaneously together, and the composite was made by splicing complete lanes together for presentation purposes. Quantification by densitometry was expressed as ratio of nuclear pFOXO1 (Ser-256) to total FOXO1 and nuclear to cytoplasmic total FOXO1. The values are presented as the means ± S.E. (n = 3). #, p < 0.05 versus Ad-NS5A control; *, p < 0.05 versus Ad-GFP control.
The effect of insulin on FOXO1 distribution in the nucleus and cytoplasm in control and NS5A protein-expressing HepG2 cells was determined in a cell fractionation study. Unlike the mRNA expression levels, the levels of nuclear FOXO1 (total) protein were similar in both the control and Ad-NS5A-infected cells. With insulin treatment, however, insulin reduced nuclear FOXO1 in the control cells, but in Ad-NS5A cells, the majority of pFOXO1 (Ser-256) was nuclear under both control and insulin treatment (Fig. 4B). These results suggest that total nuclear FOXO1 protein was not increased in NS5A cells, and although insulin increased the phosphorylated form of FOXO1 at Ser-256 in both the nuclear and cytosolic fractions, the nuclear FOXO1 remained the same in NS5A-expressing cells treated with insulin.
Impaired Proximal Insulin Signaling in Huh.8 Cells
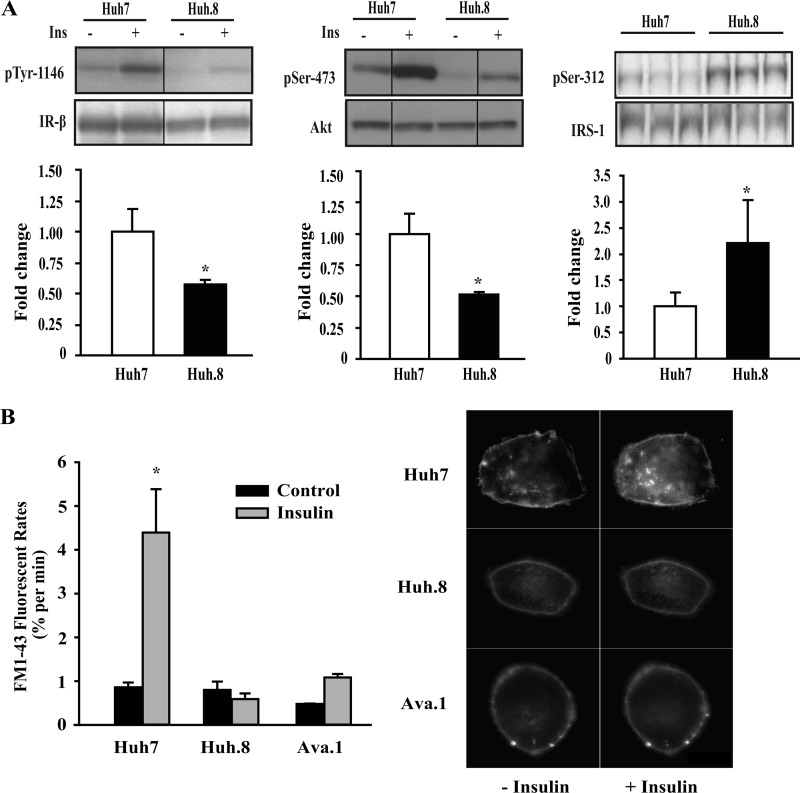
To further assess the cell signaling pathways responsible for insulin resistance in HCV-replicating cells, insulin-stimulated responses were examined in Huh7 and Huh.8 cells, as described under “Experimental Procedures.” Insulin-stimulated phosphorylation of IR-β (Tyr-1146) and phosphorylation of Akt at Ser-473 were significantly reduced by ∼50% in Huh.8 cells compared with Huh7 cells (Fig. 5A). No changes in the levels of total protein were observed with insulin treatment. Increased phosphorylation of IRS-1 (Ser-312) is an important mechanism for attenuating insulin signaling (33, 34). We observed a significant 2-fold increase in basal IRS-1 serine phosphorylation, suggesting that it may contribute to reduced Akt stimulation in HCV-expressing Huh.8 cells.
FIGURE 5.
Insulin signaling in Huh7 and Huh.8 cells. A, Huh7 and Huh.8 cells were serum-starved and treated with insulin (100 nm) or left untreated. Equivalent amounts of extracted proteins were subjected to Western blot analysis for Tyr-1146-phosphorylated IR-β relative to total IR-β, Ser-473-phosphorylated Akt relative to total Akt, and basal Ser-312-phosphorylated IRS-1 relative to total IRS-1. Representative blots are presented (for IR-β and AKT, these are composites of a larger blot where samples were run simultaneously). Insulin-stimulated fold induction relative to Huh7 (set to 1) is shown in bar graphs for pIR-β (Tyr-1146) and pAkt (Ser-473), normalized to total IR-β and Akt, respectively; and basal fold change relative to Huh7 is shown for pIRS-1 (Ser-312) normalized to total IRS-1. The values are the means ± S.E. *, p < 0.05 (n = 3). B, inhibition of the cellular response to insulin in HCV-replicating cells was measured by fluorescence assay. Mobilization of insulin-responsive vesicles was assessed by real time imaging using FM1-43 fluorescence. The rate of FM1-43 fluorescence provides a direct measurement of the cellular exocytotic activity. The dark bars represent the rates of constitutive exocytosis, and the light bars represent the rates of exocytosis after stimulation with insulin. The values are the means ± S.E. *, p < 0.05 (n = 3). Representative Huh7, Huh.8, and Ava.1 cells are shown with and without insulin treatment.
Defective Mobilization of Insulin-dependent Exocytotic Vesicles in Cells Containing HCV Subgenome Replicons
To further assess the effects of insulin, we evaluated insulin-dependent vesicular exocytosis. In liver cells, the response to insulin involves mobilization of a pool of vesicles to the plasma membrane through a PI3K-dependent pathway (28). These vesicles are sufficient in number to replace 20–30% of plasma membrane area within minutes. To evaluate whether HCV and NS5A expression has an effect on insulin-dependent exocytosis, membrane turnover was assessed in single cells using FM1-43 fluorescent dye. Because the fluorescence intensity of FM1-43 increases 350-fold with binding to biological membranes (without crossing lipid bilayers), insertion of new membrane from exocytotic vesicles is associated with a proportional increase in cellular fluorescence. In all cells, there was a gradual increase in fluorescence over time, indicating that constitutive exocytosis is sufficient to replace ∼1% of the plasma membrane every minute. Exposure to insulin in control Huh7 cells was associated with a 4-fold increase (p < 0.05) in the plasma membrane fluorescence (Fig. 5B). However, this response was completely suppressed in HCV subgenomic replicon-expressing Huh.8 cells, where insulin failed to increase the rate of exocytosis above constitutive levels. Thus, rapid changes in membrane composition in the presence of insulin were absent in HCV-replicating cells. Ava.1 cells also showed a blunted metabolic response to insulin. These findings further support the hypothesis that HCV replication and the NS5A protein causes insulin resistance in the proximal insulin signaling cascade affecting multiple pathways in these cells.
Involvement of CREB Function in HCV-expressing Cells
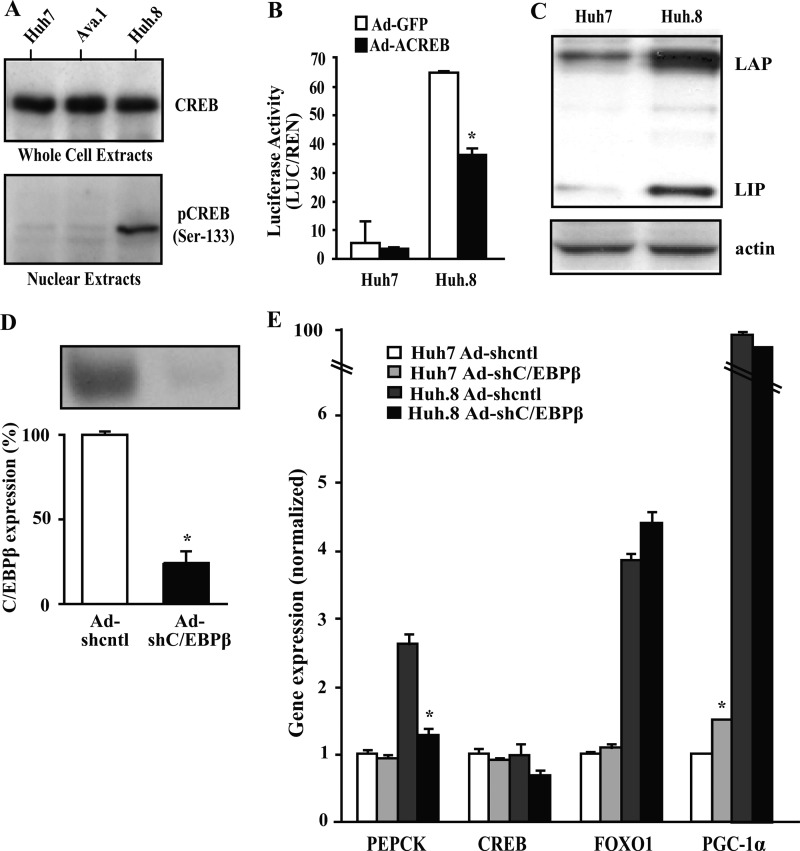
It is widely acknowledged that the cAMP signaling pathway leads to phosphorylation of CREB protein, resulting in recruitment of the coactivator CREB-binding protein p300 and subsequent activation of PEPCK and gluconeogenesis (13). Phosphorylation of CREB at Ser-133 is known to be critical for activation of CREB and binding to the PEPCK promoter, contributing to increased gluconeogenesis (13, 35, 36). To corroborate the PEPCK promoter activity data involving the CRE site in Huh.8 cells, total CREB and pCREB (Ser-133) were measured in Huh7, Huh.8, and Ava.1 cells (Fig. 6A). Although the expression of endogenous CREB protein was unchanged in all three cell lines, pCREB was increased in HCV-replicating Huh.8 cells. The mutant Ava.1 cell line lacking 47 amino acids in NS5A showed no pCREB activation, suggesting a role for full-length NS5A in the activation process.
FIGURE 6.
Involvement of CREB and C/EBPβ in HCV-mediated activation of PEPCK transcription. A, equal amounts of protein extracts (whole cell or nuclear) were collected from Huh7, Huh.8, and Ava.1 cell lines and subjected to Western blot analysis with CREB or pCREB (Ser-133). Representative blots are shown. B, Huh7 and Huh.8 cells were infected with recombinant adenovirus encoding dominant-negative CREB (Ad-ACREB) or Ad-GFP control and transfected with PEPCK-LUC. Luciferase activities of PEPCK-LUC were then measured and compared in both cell lines. The values are presented as the means ± S.E. *, p < 0.05 (n = 3). C, equivalent amounts of nuclear protein extract were collected from Huh7 and Huh.8 cell lines and subjected to Western blot analysis with C/EBPβ. Representative blots are presented. D, Huh.8 cells were infected with nontargeted or C/EBPβ shRNA adenoviruses, and expression of C/EBPβ was analyzed by Western blot. The values are presented as the means ± S.E. *, p < 0.05 (n = 2). E, comparison of transcription factor gene expression under C/EBPβ knockdown conditions in Huh7 and Huh.8 cells. RNA expression was analyzed by qPCR in Huh7 and Huh.8 cells, and the data was normalized to RPL13A expression. The values are presented as the means ± S.E. *, p < 0.05 Adcntl versus AdC/EBPβ (n = 3–5).
To further confirm a role for CREB in induction of the PEPCK promoter in Huh.8 cells, we examined PEPCK promoter activity after infection of the cells with a dominant-negative CREB adenovirus (Ad-ACREB) or control virus (Ad-GFP). The functional effectiveness of inhibiting CREB was verified in Huh.8 cells by a 70% reduction in CREB target gene Bcl-2 protein expression (data not shown). Ad-ACREB significantly reduced PEPCK promoter activity in Huh.8 cells by ∼45% (Fig. 6B). These results suggest that pCREB is an important component, but not necessarily the only activator involved in PEPCK gene expression in HCV-replicating cells.
Role of C/EBPβ in Gluconeogenic Gene Expression in Huh.8 Cells
In addition to pCREB, the transcription factor C/EBPβ also plays an important role in PEPCK gene transcription and gluconeogenesis in liver through the CRE element (37, 38). Liver expresses two forms of C/EBPβ: LAP (liver-enriched transcriptional activator protein) and a dominant-negative form, LIP (liver-enriched transcriptional inhibitory protein). Huh.8 cells expressed a 3-fold increase in LAP and LIP isoforms (Fig. 6C). To determine whether increased C/EBPβ regulates PEPCK expression in HCV-expressing cells, we infected Huh7 and Huh.8 cells with C/EBPβ or nontargeting shRNA adenoviruses. C/EBPβ knockdown significantly reduced PEPCK expression in Huh.8 cells (Fig. 6, D and E). Notably, PEPCK was normalized in Huh.8 cells treated with C/EBPβ shRNA without altering levels of CREB, FOXO1, or PGC-1α gene expression.
Increased Intracellular Lipids in HCV Replicons
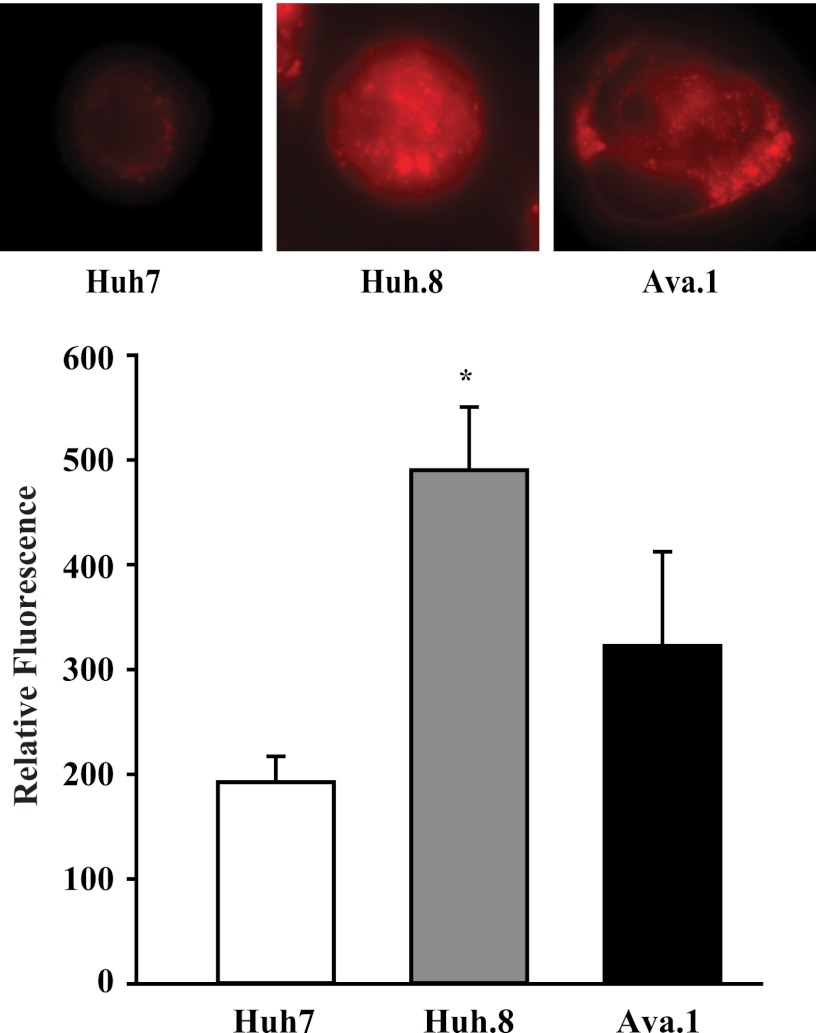
To evaluate whether the components for HCV-induced insulin resistance were paralleled by alterations in cellular fatty acid accumulation, intracellular lipids were assessed by Nile Red staining (Fig. 7). The HCV subgenome replication virus was associated with a 2.5-fold increase in relative fluorescence in Huh.8 cells compared with parental Huh7 cells, indicating an increase in intracellular lipids. The Ava.1 NS5A mutant cells showed significantly lower lipid content compared with Huh.8 cells, suggesting that expression of intact NS5A contributes to excess lipid accumulation.
FIGURE 7.
Intracellular lipids in Huh.8 cells. Huh7, Huh.8, and Ava.1 cells were stained with Nile Red, and representative cells are shown. Quantitative measurement of lipids in Huh7, Huh.8, and Ava.1 cells are shown as relative fluorescence. The values are the means ± S.E. *, p < 0.05 versus Huh7 and Ava.1 cells (n = 3).
Comparison of Lipogenic Gene Expression in Huh7 and Huh.8 Cells with C/EBPβ Knockdown
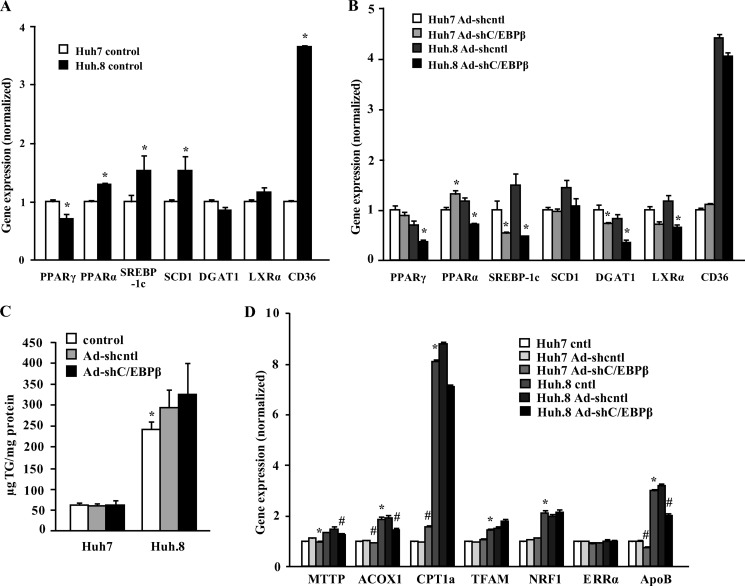
Liver TG accumulation and inflammation are pivotal metabolic hallmarks in the natural history of HCV infection, which are at least in part controlled by peroxisome proliferator-activated receptors (PPARs) and the key transcription factor sterol regulatory element-binding protein-1c (SREBP-1c) (39). We previously showed that C/EBPβ deletion prevented fatty liver and down-regulated genes that regulate hepatic lipogenesis and TG metabolism in genetically obese db/db mice and in a dietary-induced fatty liver model (17, 18). Thus, we assessed key genes involved in the lipogenic pathway in Huh7 and Huh.8 cells and whether C/EBPβ RNAi knockdown affected expression of these genes. Huh.8 cells showed a significant reduction in PPARγ together with increased PPARα expression compared with Huh7 cells (Fig. 8A). However, SREBP-1c was significantly increased in Huh.8 cells along with steroyl-CoA desaturase 1, an enzyme induced by SREBP-1c that catalyzes the synthesis of mono-unsaturated fatty acids (40). The gene for acyl CoA diacylglycerol acyltransferase 1 (DGAT1), a key enzyme that catalyzes the final step of fatty acid synthesis and is involved in fatty liver development (41), was unchanged. Liver X receptor α (LXRα), a nuclear receptor that interacts with PPARs and is involved in hepatic fatty acid synthesis, and steatosis (42), was unchanged in Huh.8 compared with Huh7 cells. The mRNA levels of the fatty acid transporter CD36 were significantly increased by 4-fold in Huh.8 cells, suggesting an increased capacity perhaps for fatty acid uptake in Huh.8 cells. Importantly, C/EBPβ knockdown in Huh.8 cells had a powerful effect to significantly suppress mRNA for PPARγ, PPARα, SREBP-1c, DGAT1, and LXRα (Fig. 8B). However, as shown in Fig. 8C, C/EBPβ knockdown in Huh.8 cells did not significantly affect TG accumulation in Huh.8 cells.
FIGURE 8.
Comparison of gene expression and TG accumulation in Huh7 and Huh.8 cells and the role of C/EBPβ knockdown. A, lipogenic gene expression was analyzed by qPCR in Huh7 and Huh.8 cells, and the data were normalized to RPL13A expression. The values are presented as the means ± S.E. *, p < 0.05 (n = 3–5). B, identical lipogenic gene expression was measured in Huh7 and Huh.8 cells after infection with nontargeting or C/EBPβ shRNA adenoviruses. The values are presented as the means ± S.E. *, p < 0.05 versus Ad-shcntl (n = 3–5). C, Huh7 and Huh.8 cells were infected with nontargeting or AdC/EBPβ shRNA adenoviruses, and TG accumulation was measured as detailed under “Experimental Procedures.” The values are presented as the means ± S.E. *, p < 0.05 versus Huh7 control (n = 2). D, fatty acid oxidation and mitochondrial transcription factor gene expression was analyzed by qPCR in Huh7 and Huh.8 cells as in A and B, and the data were normalized to RPL13A expression. The values are presented as the means ± S.E. *, p < 0.05 versus Huh7 cntl; #, p < 0.05 versus Ad-shcntl (n = 3–5).
We also investigated fatty acid oxidation gene expression and mitochondrial transcription factors in Huh7 and Huh.8 cells. Consistent with the increase in PPARα and the mitochondrial activator PGC-1α (Fig. 3), ACOX1, CPT1a, TFAM, and NRF-1 were all significantly increased in the Huh.8 cells, suggesting that reduced fatty acid oxidation is unlikely to contribute to accumulation of lipid in these cells (Fig. 8D). Apolipoprotein B, the main structural component of atherogenic lipid particles obligatory for the secretion of VLDL from the liver (43), was also increased in Huh.8 cells and was significantly reduced after C/EBPβ knockdown in both cell lines.
DISCUSSION
Epidemiological data in humans indicate that HCV infection is a strong risk for development of insulin resistance, and ultimately, overt type 2 diabetes (7). In transgenic mice expressing the HCV core gene, 100% of the mice develop hepatic insulin resistance and overt diabetes within 8 weeks, associated with an increase in TNFα in the liver (44). Our findings demonstrate that transcription of one of the first committed steps in gluconeogenesis, PEPCK and associated transcription factors, is dramatically up-regulated in Huh.8 cells, which stably express an HCV subgenome replicon, in the absence of inflammatory signals. HCV induced an increase in activation of the major gluconeogenic transcription factors pCREB (Ser-133), C/EBPβ, FOXO1, and PGC-1α. Maximal induction of PEPCK involved activation of the CRE in the PEPCK promoter, whereas infection with recombinant adenovirus encoding dominant-negative CREB or C/EBPβ RNAi in Huh.8 cells significantly reduced or normalized PEPCK expression, with no change in PGC-1α or FOXO1 levels. In addition, we demonstrate for the first time that NS5A strongly supported PEPCK gene expression and increased hepatic glucose production in HepG2 cells, whereas in Ava.1 cells with a mutation in NS5A, PEPCK expression was significantly reduced. Interestingly, although mRNA for FOXO1 was dramatically up-regulated in Huh.8 cells, neither FOXO1 protein nor the phosphorylation of FOXO1 at Ser-256 was increased in NS5A-expressing cells. Under basal conditions, Ad-NS5A infection did not change the total amount of nuclear FOXO1 but appears to increase (but not significantly) the amount of nuclear pFOXO1. With insulin, as expected, the total amount of nuclear FOXO1 is significantly reduced in control cells and appears to do the same in Ad-NS5A cells; however, nuclear pFOXO1, as a fraction of total FOXO1, is significantly greater in Ad-NS5A cells. We speculate that increased FOXO1 phosphorylation and nuclear localization in Ad-NS5A cells may be caused by a number of factors, such as decreased degradation of FOXO1 or decreased activity of the phosphatase responsible for dephosphorylation. Indeed, nuclear JNK phosphorylation is increased in HCV-infected cells (23). JNK is known to phosphorylate the 14-3-3 protein and FOXO1 at sites that prevent 14-3-3 protein binding and subsequent nuclear exportation and degradation (45). This is also consistent with the increase in the nuclear:cytostol ratio of FOXO1 seen by Deng et al. (46).
The identification of the CRE-driven gluconeogenic gene expression suggests that NS5A may play an important role in PEPCK gene expression, possibly through modulating C/EBPβ and pCREB to control gluconeogenesis. The mechanism(s) whereby NS5A activates PEPCK transcription are potentially many; however, based on our results showing the CRE is required for stimulation of PEPCK and activation of CREB, C/EBPβ, and PGC-1α (all cAMP-stimulated genes), it seems likely that NS5A plays an important role in activating the cAMP pathway.
Insulin signaling is also critical for suppressing transcription from the PEPCK promoter and gluconeogenesis (47). Indeed, the incomplete effectiveness of insulin action, caused by insulin resistance, contributes to hepatic glucose production and hyperglycemia (48). We found that NS5A stimulates increased glucose production, as well as activating pCREB and PEPCK expression, and that insulin is unable to suppress PEPCK in NS5A-infected cells. The current model of metabolic insulin resistance involves activation of serine-threonine kinase cascades (including JNK, shown previously (23)) that down-regulate IRS-1-associated PI3K activity and Akt phosphorylation, leading to increased lipogenesis and gluconeogenesis, respectively (47). Our results demonstrate that HCV significantly reduced activation of Akt signaling in Huh.8 cells and a substantial loss of insulin action to stimulate cell membrane sorting in both Huh.8 and Ava.1 cells, which is a PI3K-dependent process. This was associated with reduced insulin receptor activation, reduced IR-β expression, and increased serine phosphorylation of IRS-1 in Huh.8 cells, which has been reported previously (49).
Increased IRS-1 serine activation is consistent with our previous results showing that JNK phosphorylation is increased in HCV-expressing cells (23). Increased IRS-1 serine phosphorylation has been shown to interfere with both tyrosine phosphorylation of IRS-1 (50) and the association between IRS-1 and p85α subunit of PI3K necessary for full Akt activation (51). Whether this is due to the increase in phosphorylation of IRS-1 (Ser-312) or a combination of serine sites in IRS-1 is not proven here. Our data on FOXO1 responses to insulin demonstrate a limited response in HepG2 cells. These cells did respond metabolically to insulin to suppress glucose output, despite blunted insulin signaling; however, the net glucose output is still higher than in control cells. Our results suggest that FOXO1 levels are normal in cells infected with Ad-NS5A, but the inability of insulin to drive pFOXO1 from the nucleus suggests this may be an important mechanism, along with an increase in C/EBPβ and pCREB activation for HCV-induced PEPCK gene expression, in the absence of inflammatory factors.
HCV-expressing cells accumulated excess lipids, which may additionally play a role in the insulin resistance phenotype of these cells. Lipid infusions and high fat diets can induce insulin resistance and the impairment of insulin signaling (35) in a similar manner to our results presented herein. Interestingly, mutant NS5A reduced, but did not normalize, lipid accumulation in the context of HCV-replicating cells, suggesting other components participate in the process. HCV core proteins increase hepatic lipid accumulation via the activation of PPARγ and SREBP-1c (52) and also NS5A may exploit multiple strategies that enhance PPARγ-induced lipid accumulation (53). Our study showed a high lipid content in Huh.8 cells despite low PPARγ and increased PPARα expression. There is a similar report supporting our result with genotype 3 HCV infection (54). On the other hand, HCV increased expression of the key gene coding for the lipogenic transcription factor SREBP-1c and the fatty acid transport protein CD36, whereas LXRα (a transcriptional activator of SREBP-1c promoter) was not different, indicating a selective transcriptional regulation of lipid uptake and cholesterol synthesis as suggested previously (52, 55, 56). Interestingly, PPARγ was decreased quite significantly. However, this is consistent with a recent human study in which the intrahepatic level of PPARγ mRNA was significantly decreased in the presence of steatosis and HCV, as compared with livers without steatosis (54). When the patients were divided according to HCV genotype, PPARγ mRNA was reduced only in steatotic livers from patients infected with genotype 1 and not in those with genotype 3. PPARγ mRNA levels were invariably low, and this was independent of the presence or absence of steatosis (54). Because Huh.8 cells contain genotype 1a, our results correspond with this published data.
Huh.8 cells showed significant lipid accumulation, despite clear suppression of C/EBPβ, PPARγ, SREBP1c, and LXRα, and increased mitochondrial genes ACOX, TFAM, and Cpt1a for fatty acid oxidation. Unlike gluconeogenesis, the relative roles of various factors in the accumulation of lipid are difficult to estimate, but the net result was steatotic hepatocytes. HCV is known to inhibit lipoprotein export while increasing lipid uptake and VLDL biosynthesis (39). Currently, it is believed that HCV co-opts the VLDL assembly, maturation, degradation, and secretory machinery of the hepatocyte (reviewed in Ref. 57) to its advantage, but the finer details of this interaction are currently unclear (58). Our results show that ApoB mRNA is elevated in the Huh.8 cells and remains higher after C/EBPβ knockdown. This reinforces, albeit indirectly, that increased ApoB-VLDL assembly and/or decreased secretion may be an important part of the mechanism for increased HCV-induced steatosis. Likewise, HCV may also negatively regulate transcription of genes for mitochondrial fatty acid oxidation (57) that may contribute to retention of TGs in Huh.8 cells with C/EBPβ knockdown.
In summary, these studies demonstrate that increased PEPCK expression in HCV-replicating cells may be due to a combination of mechanisms that include loss of insulin signaling, increased CREB phosphorylation by NS5A, and an overall synergistic activation by modification of factors binding to the CRE, as well as the GRE sites on the PEPCK promoter. The data presented here clearly show that the cAMP-driven pathways via C/EBPβ and pCREB are highly active in HCV, but that NS5A is an important viral component in PEPCK gene expression and lipid retention in HCV-replicating cells. The retention of insulin resistance in Ava.1 cells suggests that viral particles in addition to NS5A may contribute to the mechanism(s) for HCV-induced insulin resistance associated with HCV infection. Given that PEPCK has a central role in regulation of gluconeogenesis and the progression to diabetes, these observations suggest further defining the cellular mechanisms responsible for insulin resistance and gluconeogenic gene expression by NS5A, and C/EBPβ represents an important new focus for understanding a cellular basis for the metabolic complications during HCV infection that may lead to improved clinical outcomes.
Acknowledgment
We thank Dr. Charles Rice (Laboratory of Virology and Infectious Disease, Center for the Study of Hepatitis C, The Rockefeller University) for providing the Huh7, Huh.8, and Ava.1 cell lines.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK46802 (to G. K.), R01-DK43278 (to J. G. F.), R01-DK46082 (to J. G. F.), and R01-DK59767 (to J. E. F.). This work was also supported by funding from the Herman Lopeta Memorial Award and the American Liver Foundation (to I. Q.) and by an American Diabetes Association Mentored Post-Doctoral Fellowship (to M. C.).
- HCV
- hepatitis C virus
- IRS-1
- insulin receptor substrate
- NS5A
- nonstructural component 5A
- PEPCK
- phosphoenolpyruvate carboxykinase
- FOXO1
- forkhead box protein O1
- CREB
- cAMP response element-binding protein
- C/EBP
- CCAAT/enhancer-binding protein
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1
- G-6-Pase
- glucose-6-phosphatase
- Ad
- adenovirus
- GRE
- glucocorticoid response element
- CRE
- cAMP response element
- IR-β
- insulin receptor β
- qPCR
- quantitative real time PCR
- TG
- triglyceride
- PPAR
- peroxisome proliferator-activated receptor
- SREBP-1c
- sterol regulatory element-binding protein-1c
- LXRα
- liver X receptor α.
REFERENCES
- 1. Ray Kim W. (2002) Global epidemiology and burden of hepatitis C. Microbes Infect. 4, 1219–1225 [DOI] [PubMed] [Google Scholar]
- 2. Monto A., Alonzo J., Watson J. J., Grunfeld C., Wright T. L. (2002) Steatosis in chronic hepatitis C. Relative contributions of obesity, diabetes mellitus, and alcohol. Hepatology 36, 729–736 [DOI] [PubMed] [Google Scholar]
- 3. Ortiz V., Berenguer M., Rayón J. M., Carrasco D., Berenguer J. (2002) Contribution of obesity to hepatitis C-related fibrosis progression. Am. J. Gastroenterol. 97, 2408–2414 [DOI] [PubMed] [Google Scholar]
- 4. Thuluvath P. J., John P. R. (2003) Association between hepatitis C, diabetes mellitus, and race. A case-control study. Am. J. Gastroenterol. 98, 438–441 [DOI] [PubMed] [Google Scholar]
- 5. Wang C. S., Wang S. T., Yao W. J., Chang T. T., Chou P. (2003) Community-based study of hepatitis C virus infection and type 2 diabetes. An association affected by age and hepatitis severity status. Am. J. Epidemiol. 158, 1154–1160 [DOI] [PubMed] [Google Scholar]
- 6. Hui J. M., Sud A., Farrell G. C., Bandara P., Byth K., Kench J. G., McCaughan G. W., George J. (2003) Insulin resistance is associated with chronic hepatitis C virus infection and fibrosis progression [corrected]. Gastroenterology 125, 1695–1704 [DOI] [PubMed] [Google Scholar]
- 7. Kawaguchi T., Sata M. (2010) Importance of hepatitis C virus-associated insulin resistance. Therapeutic strategies for insulin sensitization. World J. Gastroenterol. 16, 1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aytug S., Reich D., Sapiro L. E., Bernstein D., Begum N. (2003) Impaired IRS-1/PI3-kinase signaling in patients with HCV. A mechanism for increased prevalence of type 2 diabetes. Hepatology 38, 1384–1392 [DOI] [PubMed] [Google Scholar]
- 9. Lerat H., Honda M., Beard M. R., Loesch K., Sun J., Yang Y., Okuda M., Gosert R., Xiao S. Y., Weinman S. A., Lemon S. M. (2002) Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122, 352–365 [DOI] [PubMed] [Google Scholar]
- 10. Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Ishibashi K., Matsuura Y., Miyamura T., Koike K. (1997) Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78, 1527–1531 [DOI] [PubMed] [Google Scholar]
- 11. Kawaguchi T., Yoshida T., Harada M., Hisamoto T., Nagao Y., Ide T., Taniguchi E., Kumemura H., Hanada S., Maeyama M., Baba S., Koga H., Kumashiro R., Ueno T., Ogata H., Yoshimura A., Sata M. (2004) Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am. J. Pathol. 165, 1499–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McLauchlan J. (2009) Hepatitis C virus. Viral proteins on the move. Biochem. Soc. Trans. 37, 986–990 [DOI] [PubMed] [Google Scholar]
- 13. Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 [DOI] [PubMed] [Google Scholar]
- 14. Kamagate A., Qu S., Perdomo G., Su D., Kim D. H., Slusher S., Meseck M., Dong H. H. (2008) FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J. Clin. Invest. 118, 2347–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsumoto M., Pocai A., Rossetti L., Depinho R. A., Accili D. (2007) Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 [DOI] [PubMed] [Google Scholar]
- 16. Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 17. Rahman S. M., Schroeder-Gloeckler J. M., Janssen R. C., Jiang H., Qadri I., Maclean K. N., Friedman J. E. (2007) CCAAT/enhancing binding protein β deletion in mice attenuates inflammation, endoplasmic reticulum stress, and lipid accumulation in diet-induced nonalcoholic steatohepatitis. Hepatology 45, 1108–1117 [DOI] [PubMed] [Google Scholar]
- 18. Schroeder-Gloeckler J. M., Rahman S. M., Janssen R. C., Qiao L., Shao J., Roper M., Fischer S. J., Lowe E., Orlicky D. J., McManaman J. L., Palmer C., Gitomer W. L., Huang W., O'Doherty R. M., Becker T. C., Klemm D. J., Jensen D. R., Pulawa L. K., Eckel R. H., Friedman J. E. (2007) CCAAT/enhancer-binding protein β deletion reduces adiposity, hepatic steatosis, and diabetes in Lepr(db/db) mice. J. Biol. Chem. 282, 15717–15729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rahman S. M., Qadri I., Janssen R. C., Friedman J. E. (2009) Fenofibrate and PBA prevent fatty acid-induced loss of adiponectin receptor and pAMPK in human hepatoma cells and in hepatitis C virus-induced steatosis. J. Lipid Res. 50, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blight K. J., Kolykhalov A. A., Rice C. M. (2000) Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974 [DOI] [PubMed] [Google Scholar]
- 21. Tellinghuisen T. L., Paulson M. S., Rice C. M. (2006) The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein. J. Virol. 80, 7450–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choudhury M., Shukla S. D. (2008) Surrogate alcohols and their metabolites modify histone H3 acetylation. Involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin. Exp. Res. 32, 829–839 [DOI] [PubMed] [Google Scholar]
- 23. Qadri I., Iwahashi M., Capasso J. M., Hopken M. W., Flores S., Schaack J., Simon F. R. (2004) Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication. Role of JNK, p38 MAPK and AP-1. Biochem. J. 378, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao J., Qiao L., Janssen R. C., Pagliassotti M., Friedman J. E. (2005) Chronic hyperglycemia enhances PEPCK gene expression and hepatocellular glucose production via elevated liver activating protein/liver inhibitory protein ratio. Diabetes 54, 976–984 [DOI] [PubMed] [Google Scholar]
- 25. Routes J. M., Colton L. A., Ryan S., Klemm D. J. (2000) CREB (cAMP response element binding protein) and C/EBPα (CCAAT/enhancer binding protein) are required for the superstimulation of phosphoenolpyruvate carboxykinase gene transcription by adenoviral E1a and cAMP. Biochem. J. 352, 335–342 [PMC free article] [PubMed] [Google Scholar]
- 26. Rahman S. M., Janssen R. C., Choudhury M., Baquero K. C., Aikens R. M., de la Houssaye B. A., Friedman J. E. (2012) CCAAT/enhancer binding protein β (C/EBPβ) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perdomo G., Commerford S. R., Richard A. M., Adams S. H., Corkey B. E., O'Doherty R. M., Brown N. F. (2004) Increased β-oxidation in muscle cells enhances insulin-stimulated glucose metabolism and protects against fatty acid-induced insulin resistance despite intramyocellular lipid accumulation. J. Biol. Chem. 279, 27177–27186 [DOI] [PubMed] [Google Scholar]
- 28. Kilic G., Doctor R. B., Fitz J. G. (2001) Insulin stimulates membrane conductance in a liver cell line. Evidence for insertion of ion channels through a phosphoinositide 3-kinase-dependent mechanism. J. Biol. Chem. 276, 26762–26768 [DOI] [PubMed] [Google Scholar]
- 29. Chakravarty K., Cassuto H., Reshef L., Hanson R. W. (2005) Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit. Rev. Biochem. Mol. Biol. 40, 129–154 [DOI] [PubMed] [Google Scholar]
- 30. Hanson R. W., Reshef L. (1997) Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 66, 581–611 [DOI] [PubMed] [Google Scholar]
- 31. Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 [DOI] [PubMed] [Google Scholar]
- 32. Shoji I., Deng L., Hotta H. (2011) Molecular mechanism of hepatitis C virus-induced glucose metabolic disorders. Front. Microbiol. 2, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aguirre V., Werner E. D., Giraud J., Lee Y. H., Shoelson S. E., White M. F. (2002) Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J. Biol. Chem. 277, 1531–1537 [DOI] [PubMed] [Google Scholar]
- 34. Tanti J. F., Gual P., Grémeaux T., Gonzalez T., Barres R., Le Marchand-Brustel Y. (2004) Alteration in insulin action. Role of IRS-1 serine phosphorylation in the retroregulation of insulin signalling. Ann. Endocrinol. (Paris) 65, 43–48 [DOI] [PubMed] [Google Scholar]
- 35. Shulman G. I. (2000) Cellular mechanisms of insulin resistance. J. Clin. Invest. 106, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bevan P. (2001) Insulin signalling. J. Cell Sci. 114, 1429–1430 [DOI] [PubMed] [Google Scholar]
- 37. Arizmendi C., Liu S., Croniger C., Poli V., Friedman J. E. (1999) The transcription factor CCAAT/enhancer-binding protein β regulates gluconeogenesis and phosphoenolpyruvate carboxykinase (GTP) gene transcription during diabetes. J. Biol. Chem. 274, 13033–13040 [DOI] [PubMed] [Google Scholar]
- 38. Park E. A., Gurney A. L., Nizielski S. E., Hakimi P., Cao Z., Moorman A., Hanson R. W. (1993) Relative roles of CCAAT/enhancer-binding protein β and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP). J. Biol. Chem. 268, 613–619 [PubMed] [Google Scholar]
- 39. Lerat H., Kammoun H. L., Hainault I., Mérour E., Higgs M. R., Callens C., Lemon S. M., Foufelle F., Pawlotsky J. M. (2009) Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J. Biol. Chem. 284, 33466–33474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oshino N., Imai Y., Sato R. (1966) Electron-transfer mechanism associated with fatty acid desaturation catalyzed by liver microsomes. Biochim. Biophys. Acta 128, 13–27 [DOI] [PubMed] [Google Scholar]
- 41. Villanueva C. J., Monetti M., Shih M., Zhou P., Watkins S. M., Bhanot S., Farese R. V., Jr. (2009) Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology 50, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hong C., Tontonoz P. (2008) Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr. Opin. Genet. Dev. 18, 461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis R. A. (1999) Cell and molecular biology of the assembly and secretion of apolipoprotein B-containing lipoproteins by the liver. Biochim. Biophys. Acta 1440, 1–31 [DOI] [PubMed] [Google Scholar]
- 44. Shintani Y., Fujie H., Miyoshi H., Tsutsumi T., Tsukamoto K., Kimura S., Moriya K., Koike K. (2004) Hepatitis C virus infection and diabetes. Direct involvement of the virus in the development of insulin resistance. Gastroenterology 126, 840–848 [DOI] [PubMed] [Google Scholar]
- 45. Singh A., Ye M., Bucur O., Zhu S., Tanya Santos M., Rabinovitz I., Wei W., Gao D., Hahn W. C., Khosravi-Far R. (2010) Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol. Biol. Cell 21, 1140–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deng L., Shoji I., Ogawa W., Kaneda S., Soga T., Jiang D. P., Ide Y. H., Hotta H. (2011) Hepatitis C virus infection promotes hepatic gluconeogenesis through an NS5A-mediated, FoxO1-dependent pathway. J. Virol. 85, 8556–8568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barthel A., Schmoll D. (2003) Novel concepts in insulin regulation of hepatic gluconeogenesis. Am. J. Physiol. Endocrinol. Metab. 285, E685–E692 [DOI] [PubMed] [Google Scholar]
- 48. Davis R. C., Castellani L. W., Hosseini M., Ben-Zeev O., Mao H. Z., Weinstein M. M., Jung D. Y., Jun J. Y., Kim J. K., Lusis A. J., Péterfy M. (2010) Early hepatic insulin resistance precedes the onset of diabetes in obese C57BLKS-db/db mice. Diabetes 59, 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Banerjee S., Saito K., Ait-Goughoulte M., Meyer K., Ray R. B., Ray R. (2008) Hepatitis C virus core protein upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream akt/protein kinase B signaling pathway for insulin resistance. J. Virol. 82, 2606–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu C., Chen Y., Cline G. W., Zhang D., Zong H., Wang Y., Bergeron R., Kim J. K., Cushman S. W., Cooney G. J., Atcheson B., White M. F., Kraegen E. W., Shulman G. I. (2002) Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 277, 50230–50236 [DOI] [PubMed] [Google Scholar]
- 51. Weigert C., Kron M., Kalbacher H., Pohl A. K., Runge H., Häring H. U., Schleicher E., Lehmann R. (2008) Interplay and effects of temporal changes in the phosphorylation state of serine-302, -307, and -318 of insulin receptor substrate-1 on insulin action in skeletal muscle cells. Mol. Endocrinol. 22, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim K. H., Hong S. P., Kim K., Park M. J., Kim K. J., Cheong J. (2007) HCV core protein induces hepatic lipid accumulation by activating SREBP1 and PPARγ. Biochem. Biophys. Res. Commun. 355, 883–888 [DOI] [PubMed] [Google Scholar]
- 53. Kim K., Kim K. H., Ha E., Park J. Y., Sakamoto N., Cheong J. (2009) Hepatitis C virus NS5A protein increases hepatic lipid accumulation via induction of activation and expression of PPARγ. FEBS Lett. 583, 2720–2726 [DOI] [PubMed] [Google Scholar]
- 54. de Gottardi A., Pazienza V., Pugnale P., Bruttin F., Rubbia-Brandt L., Juge-Aubry C. E., Meier C. A., Hadengue A., Negro F. (2006) Peroxisome proliferator-activated receptor-α and -γ mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment Pharmacol. Ther. 23, 107–114 [DOI] [PubMed] [Google Scholar]
- 55. Park C. Y., Jun H. J., Wakita T., Cheong J. H., Hwang S. B. (2009) Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J. Biol. Chem. 284, 9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackel-Cram C., Qiao L., Xiang Z., Brownlie R., Zhou Y., Babiuk L., Liu Q. (2010) Hepatitis C virus genotype-3a core protein enhances sterol regulatory element-binding protein-1 activity through the phosphoinositide 3-kinase-Akt-2 pathway. J. Gen. Virol. 91, 1388–1395 [DOI] [PubMed] [Google Scholar]
- 57. Syed G. H., Amako Y., Siddiqui A. (2010) Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol. Metab. 21, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bugianesi E., Salamone F., Negro F. (2012) The interaction of metabolic factors with HCV infection. Does it matter? J. Hepatol. 56, (Suppl. 1) S56–S65 [DOI] [PubMed] [Google Scholar]