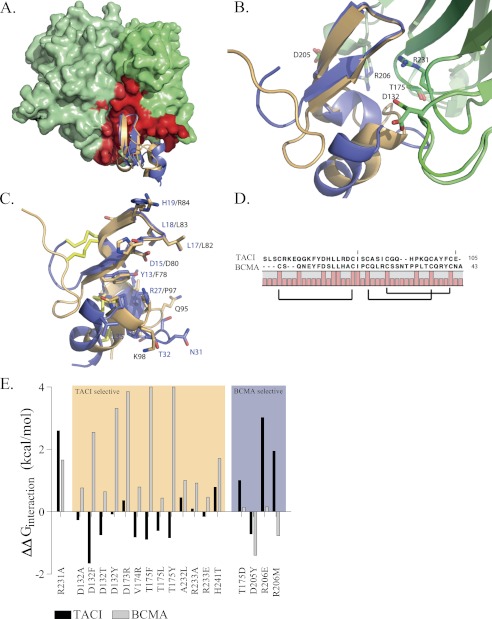
FIGURE 1.
Crystal Structures of APRIL in complex with BCMA and TACI and prediction of the APRIL-selective mutants. A, front view of APRIL (light and dark green) in complex with TACI (orange) or BCMA (blue). The APRIL-TACI and APRIL-BCMA complexes are superimposed. APRIL monomers are depicted using a molecular surface representation, and main chain coordinates of the receptors are depicted schematically. For clarity, only a single receptor unit is depicted, and two ligand monomers are shown. The TACI and BCMA receptor-binding interface of APRIL is mapped in red on the APRIL surface. In contrast to most other TNF family ligands, the receptor-binding interface resides only for a small part in the cleft between two adjacent ligand monomers because most of the receptors interactions are located on the central surface of a single APRIL monomer. B, detailed view of TACI and BCMA in complex with APRIL. Selected APRIL residues involved in an interaction with the receptors are depicted (APRIL structure in complex with BCMA and TACI is depicted in light green or dark green, respectively). C, detailed view of TACI (orange) and BCMA (blue) residues involved in APRIL binding. TACI and BCMA show a root mean square deviation of 1.45 Å upon superposition (calculated over 95 main chain atoms); the main chain coordinates show a larger displacement C-terminally of the β-sheet. The interacting residues of BCMA or TACI are relatively non-conserved. Labels of BCMA residues are colored blue, TACI is shown in black, and cysteine bridges are colored yellow. D, structure-based alignment of the ECD ligand-binding domain of human BCMA and human TACI. Brackets indicate cysteine bridge connectivity. Full bars, conserved residues. E, FoldX interaction energy. Interaction free energy between APRIL variants and BCMA or TACI is calculated as the difference with the interaction energy of wild type APRIL and expressed as ΔΔG in kcal/mol. The FoldX interaction energy is corrected for unfavorable intrachain Van der Waals clashes upon mutation (see “Experimental Procedures”). Variants are grouped as TACI-specific or BCMA-specific. R231A, a previously constructed APRIL variant unable to bind both receptors, was used as control. Structure images were generated using PyMOL (available on the World Wide Web) and based on Protein Data Bank entries 1xu1 and 1xu2 (21).