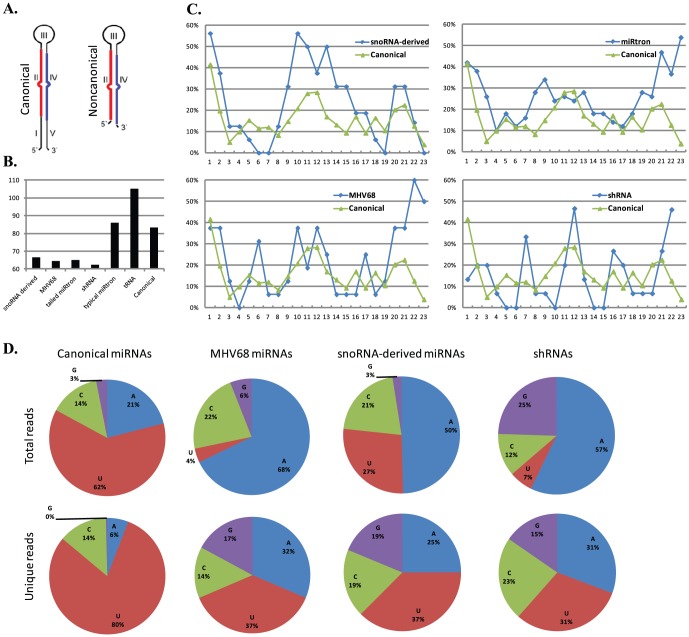
Figure 4. Features of canonical and noncanonical miRNAs.
(A) A diagram marks the locations of the five regions for canonical miRNA hairpins: I, loop-distal (5′ arm); II, miRNA-5p; III, loop-proximal; IV, miRNA-3p; V, loop-distal (3′ arm). Noncanonical miRNA hairpins miss I and V loop-distal regions. (B) The average length of hairpins for canonical miRNAs, snoRNA-, tRNA-derived miRNAs, miRtrons (typical and tailed) and MHV68-encoded miRNAs (C) The percentage of the number of unpaired bases at each position (from 5′ to 3′) for canonical (green) and noncanonical (blue) miRNAs, respectively. (D) First nucleotide percentage of total reads (upper) in MHV68 data mapped to conserved, MHV68 encoded, snoRNA-derived miRNAs and shRNAs. Percentages of unique miRNAs are presented in the lower part, respectively.