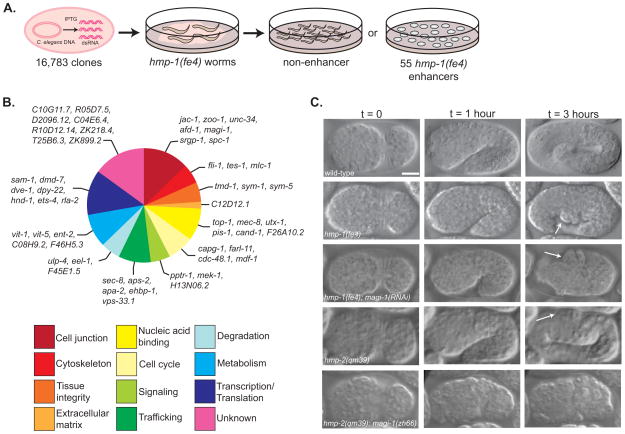
Figure 1. An enhancer screen identifies putative adhesion modulators during C. elegans morphogenesis.
(A) We screened a library of 16,783 RNAi feeding clones to identify enhancers of a hypomorphic allele of α-catenin, hmp-1(fe4). Each bacterial clone was fed to wild-type and hmp-1(fe4) worms and phenotypes were assessed in the next generation. Enhancers increased lethality in hmp-1(fe4) embryos to >83% while causing <10% lethality in wild-type embryos. (B) Putative functions for enhancers were determined using literature searches, BLAST and ClustalW comparisons of protein sequence, identification of domains using Pfam, and NCBI KOG description. Functional groups were based on previously published categories [37, 38]. (C) Knocking down magi-1 enhances the penetrance and severity of hmp-1(fe4) and hmp-2(qm39) phenotypes. Wild-type embryos enclose (t = 0), turn (t = 1 hour), and elongate ~4-fold their original length before hatching (t = 3 hours). hmp-1(fe4) embryos enclose (t = 0), but fail to fully elongate, developing body shape defects (arrow, t = 3 hours). The majority of hmp-1(fe4); magi-1(RNAi) embryos enclose (t = 0), but almost immediately retract dorsally and arrest, with a characteristic humpback phenotype (arrows, t = 1 and 3 hours). hmp-2(qm39) embryos enclose (t = 0) and elongate, occasionally developing mild body shape defects (arrow, t = 3 hours). A significant fraction of hmp-2(qm39); magi-1(zh66) embryos fail to enclose and rupture (t = 0). Scale bar is 10μm.