Abstract
A process for the preparation of an abiotic protein affinity ligand is described. The affinity ligand, a synthetic polymer hydrogel nanoparticle (NP), is formulated with functional groups complementary to the surface presentation of the target protein. An iterative process is used to improve affinity by optimizing the composition and proportion of functional monomers. Since the polymer NPs are formed by a kinetically driven process, the sequence of functional monomers in the polymer chain is not controlled; only the average composition can be adjusted by the stoichiometry of the monomers in the feed. To compensate for this the hydrogel NP is lightly crosslinked resulting in chain flexibility that takes place on a sub millisecond time scale allowing the polymer to “map” onto a protein surface with complementary functionality. In this study, we report a lightly crosslinked (2%) N-isopropyl acrylamide (NIPAm) synthetic polymer NP (50~65 nm) incorporating hydrophobic and carboxylate groups, binds with high affinity to the Fc fragment of IgG. The affinity and amount of NP bound to IgG is pH dependent. The hydrogel NP inhibits protein A binding to the Fc domain at pH 5.5, but not at pH 7.3. A computational analysis was used to identify potential NP-protein interaction sites. Candidates include a NP binding domain that overlaps with the protein A-Fc binding domain at pH 5.5. The computational analysis supports the inhibition experimental results and is attributed to the difference in the charged state of histidine residues. Affinity of the NP (3.5~8.5 nM) to the Fc domain at pH 5.5 is comparable to protein A at pH 7. These results establish that engineered synthetic polymer NPs can be formulated with an intrinsic affinity to a specific domain of a large biomacromolecule.
INTRODUCTION
Nanomedicine is driven by the premise that discrete synthetic nanoparticles (NPs) can be formulated to target specific proteins, cells or organs. NP targeting coupled with function (drug delivery, imaging, diagnostics, concentration, isolation and purification) provides opportunities for transformative approaches to therapeutics, diagnostics and biomacromolecule isolation and purification. This is a vibrant area of research with recent successes that include therapeutic reagents,1,2 drug delivery vehicles,3–5 sensors,6–8 toxin neutralization9–11 and enzyme inhibition.12,13 NP specificity for target biomolecules is most often accomplished by the attachment of affinity ligands, including antibodies. The need for a comprehensive collection of affinity agents for proteins has been heightened by National Institutes of Health’s (NIH’s) broad initiative to obtain multiple capture agents for all proteins in the proteome.14
Recombinant antibodies are the current gold standard of affinity agents and it is likely they will play a dominant role for the foreseeable future. However, antibodies are not without some limitations. For example, the cost of developing new protein capture agents is high. The time required for discovery of an effective antibody can also be lengthy. Some proteins may not function for all intended applications. These and related issues create practical challenges to formulating a comprehensive set of antibody target capture reagents. In addition to antibodies, alternative technologies that include peptides, peptide mimics and aptamers offer promising opportunities to expand the candidate pool of protein capture reagents.15–17 Considering the range of targets and uses, it is likely that a combination of approaches will be needed to generate a comprehensive resource.
We have been developing an alternative approach for protein and peptide capture agents. Our strategy takes cognizance of the fact that protein-protein interaction surfaces span hundreds of square angstroms.18 Affinity arises from the cumulative effect of individually weak interactions that include van der Waals, hydrogen bonding, and electrostatic interactions. Our capture agent, a synthetic polymer hydrogel, is formulated with functional groups complementary to protein domains or peptide targets. We then use an iterative process to improve affinity to a target peptide or protein by optimizing the composition and proportion of functional monomers. Since the polymer NPs are formed by a kinetically driven process, the sequence of functional monomers in the polymer chain is not controlled; only the average composition of the polymer can be adjusted by the stoichiometry of the monomers in the feed. However, to compensate for this the hydrogel NP is lightly crosslinked (~2%) resulting in considerable chain flexibility that takes place on a sub millisecond time scale19. This allows the polymer to “map” onto a protein surface with complementary functionality compensating in part for the lack of sequence and topological control of the synthetic polymer NP.
Our previous efforts focused on synthetic polymer NPs with antibody-like affinity and selectivity to a toxic peptide, melittin. Polymer NPs with low nanomolar affinity and high selectivity were developed and were shown to function by neutralizing the peptides toxicity in vitro and in vivo.10,20 The present study describes an important step beyond peptide recognition and capture, specifically, progress in developing a synthetic polymer NP that binds to a specific targeted domain of a large protein.
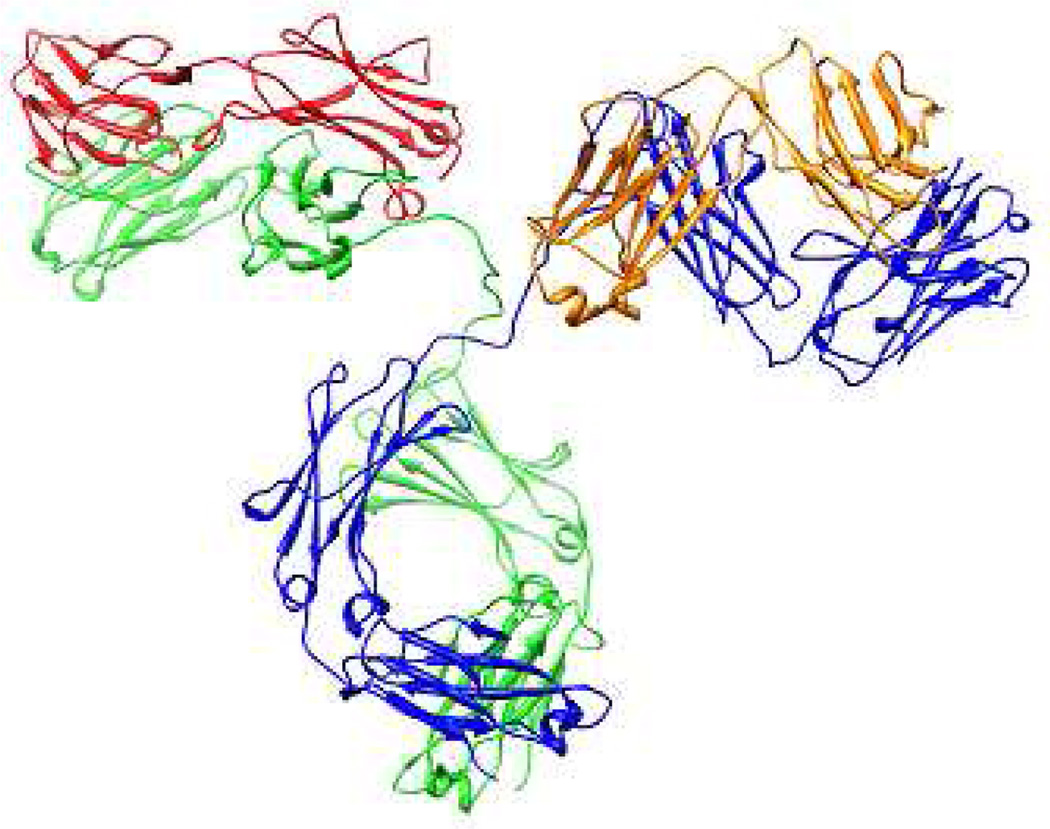
The protein target of this study is the 150 kDa protein immunoglobulin G (IgG). IgG is the workhorse protein for research, diagnostics and increasingly, therapeutic applications.21,22 IgGs are composed of 4 protein chains, 2 identical heavy chains of approximately 50 kDa and 2 identical light chains of about 25 kDa. The tetramer has two identical halves that form a forked Y shape. Each end of the fork contains an identical antigen-binding site (the variable regions). The structure is given in Figure 1, which also highlights heavy and light chains. The base or conserved region of the structure corresponds to the Fc fragment (Figure 1) and can be cleaved from IgG and studied independently. Purification of recombinant IgG from cellular extract is often achieved by a multi step process that includes chromatographic separation using protein affinity ligands that strongly and specifically bind to the conserved Fc domain. Protein A or G are the most common affinity ligands used for purification of IgG. Protein A and G themselves are products of recombinant technology and must be expressed and purified. It is not surprising therefore that considerable effort has been expended in the design of alternatives to protein A/G. These alternatives have included synthetic ligands,22,23 DNA/RNA aptamers,16 peptides,15, 17 and synthetic polymers.24,25 The utility of the Fc domain as a target for protein capture dictated its choice for our study.
Figure 1.
The structure of IgG with heavy chains in blue and green and light chains in orange and red. The Fc-Fc dimer is shown at the bottom of the image. The PDB ID is 1HZH.
A small library of multifunctional polymer NPs containing varying amounts of hydrophobic, hydrophilic and charged monomers were screened for IgG affinity and the “hits” were taken into a second round of NP synthesis, tuning the composition of critical monomers to arrive at NPs with high affinity for IgG. A more focused effort followed to establish that the dominant NP-protein interaction is with the Fc domain of IgG. This effort was coupled with analysis of the Fc protein surface to identify potential Fc-NP binding domains and their response to changes in pH. Finally a NP-protein A competition was performed to help identify the location of the dominant NP-IgG interaction.
RESULTS AND DISSCUSSION
Synthesis and screening of NP-IgG affinity
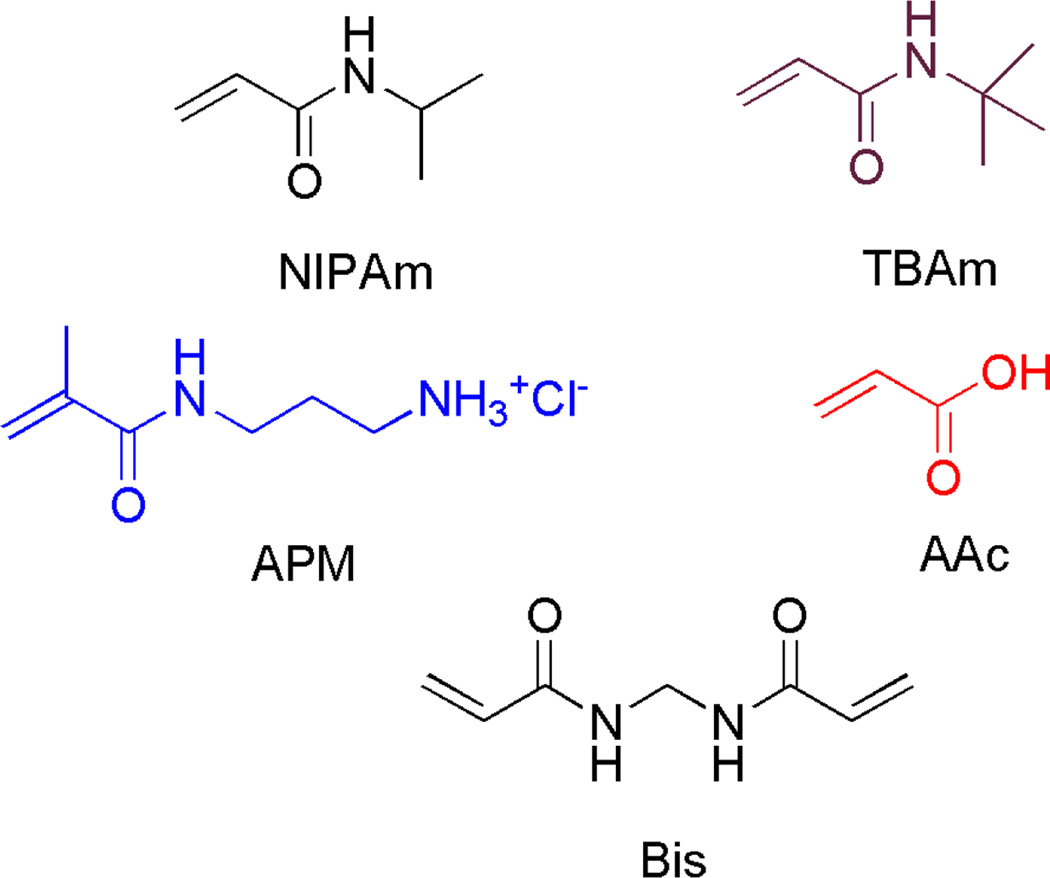
A small library of multifunctional hydrogel NPs (NP1-6) were synthesized by precipitation polymerization.10,26,27 NIPAm was the core monomer in combination with various amounts of AAc, APM, and TBAm, as negatively-charged, positively-charged and hydrophobic functional monomers respectively. Bis, 2 mol%) was incorporated as a crosslinker (Figure 2). NP size was established by dynamic light scattering (DLS, Table 1). Table S1 summarizes the feed ratio and reaction conditions used to prepare the NP library. DLS was also used in the initial screen of NP-IgG interactions in water28,29. An association between a NP and IgG (10~12 nm)28 should result in an observable increase in particle size. From this screen, NP6 incorporating both TBAm and AAc monomers showed a significant increase in size following addition of IgG (Table 1). NP1 and 4 also exhibited an increase in size upon addition of IgG.
Figure 2.
Monomers used for NP synthesis
Table 1.
Composition, yield and diameter of NPs in pure water in the absence/presence of IgG (133 nM) in water
| NP# | Feed ratio of functional monomers [%]a |
Yield [%] | Diameter [nm]b |
Diameter after the addition of IgG [nm]c |
||
|---|---|---|---|---|---|---|
| TBAm | APM | AAc | ||||
| 1 | 40 | 0 | 0 | 61.6 | 56 | Aggregation d |
| 2 | 0 | 5 | 0 | 30.1 | 94 | 89 |
| 3 | 0 | 0 | 5 | 84.1 | 20 | 20 |
| 4 | 0 | 0 | 0 | 87.7 | 88 | 105 |
| 5 | 40 | 5 | 0 | 26.4 | 110 | 110 |
| 6 | 40 | 0 | 5 | 64.0 | 60 | Aggregation d |
| 7 | 40 | 0 | 20 | 65.2 | 64 | Aggregation d |
| 8 | 40 | 0 | 40 | 48 | 58 | Aggregation d |
NIPAm is present according to the following relationship (NIPAm = 98-TBAm + APM + AAc). In all cases, Bis is present as 2% of the total monomer composition.
Average of at least 3 independent measurements (standard deviation 0.2 to 1.9 nm)
Average of at least 3 independent measurements (standard deviation 0.1 to 2.1 nm)
Aggregation is defined by the observation of poly-modality with formation of micron-sized species.
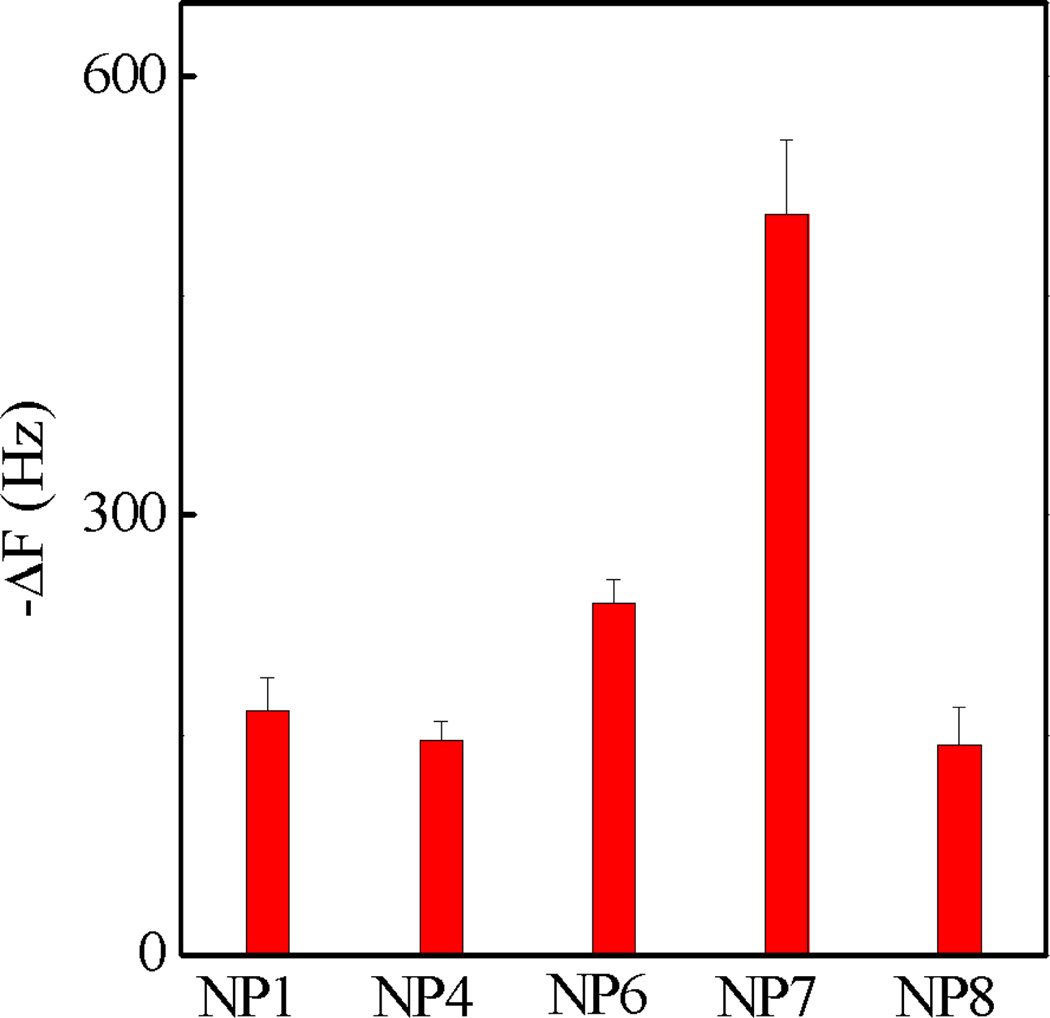
The interactions of NP1, 4 and 6 with Fc-orientated IgG were evaluated by QCM in PBS (35 mM, pH= 7.3 with 150 mM NaCl). IgG orientation was achieved by first immobilizing its antigen, TNFα, on the QCM surface. The surface was then functionalized with humanized monoclonal IgG (anti-TNFα antibody).30 Since TNFα binds the Fab fragment of IgG, this orientates the Fc domain into the bulk solution making it accessible to NPs. A 0.1% BSA solution was subsequently added to reduce non-specific binding. To confirm the Fc domain was accessible from solution, the prepared surface was exposed to Au NPs coated with an antihuman IgG which specifically binds to the Fc domain of IgG. The strong signal confirmed that the Fc domain was accessible to solution affinity ligands (Figure S1), validating the use of this method to screen NP affinity for the Fc domain of IgG. The binding of NP1, 4 and 6 to the oriented IgG is summarized in Figure 3. NP 6 showed greater frequency change than NP1 and 4. The NPs are comprised of functional groups that could participate in hydrogen bonding, hydrophobic and electrostatic interactions with proteins. This preliminary result suggests that a combination of these functional groups is necessary for binding.
Figure 3.
QCM screening of the library of polymer NPs containing 40% TBAm and varying amounts of AAc with Fc-oriented IgG in PBS (35 mM, pH 7.3 with 150 mM NaCl). Standard deviations were calculated from three independent measurements.
Optimization of NP composition and evaluation of NP binding specificity
In an effort to maximize NP affinity to IgG, an expanded pool of NPs was prepared. One series contained greater amounts of AAc than NP6 (NP7 and NP8, Table 1 and S2). However, efforts to prepare NPs with greater amounts of TBAm were not successful due to the colloidal instability of the resulting materials. The binding of NP1, 6, 7, 8 to the oriented IgG is summarized in Figure 3. The frequency change of NP7 (20% AAc, 40% TBAm) was two to three times higher than other NPs. This NP was taken on for further experiments.
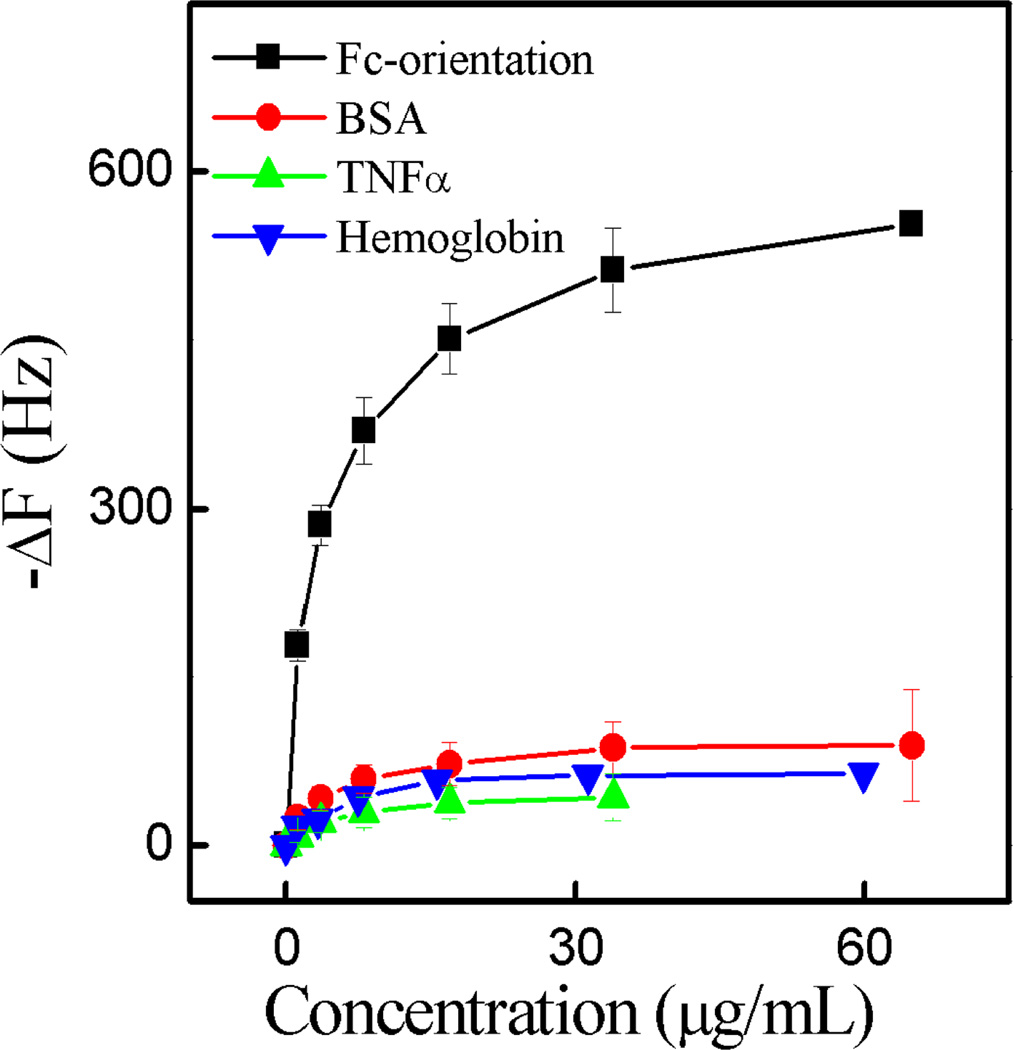
In addition to IgG affinity a preliminary screen for protein specificity was undertaken. The interaction of BSA, TNFα and hemoglobin against NP7 was studied in PBS (35 mM, pH 7.3 with 150 mM NaCl). NP7 had little interaction with these proteins (Figure 4), suggesting that in the above diagnostic, the observed NP affinity for IgG was primarily associated with the oriented Fc domain of IgG. In addition, multivalency of the interaction between NP7 and IgG was established by observing changes in particle size by DLS before and after the addition of IgG to an aqueous solution of NPs. At a molar ratio of IgG to NP of 1.25, aggregates formed (Figure S2). We interpret this observation as multivalency of both IgG and the NPs.
Figure 4.
The interactions between NP7 and IgG, BSA, TNFα and hemoglobin in each of which were immobilized on a QCM surface in PBS (35 mM, pH 7.3 with 150 mM NaCl). Standard deviations were calculated from three independent measurements.
Effect of pH and buffer on NP-Protein interactions
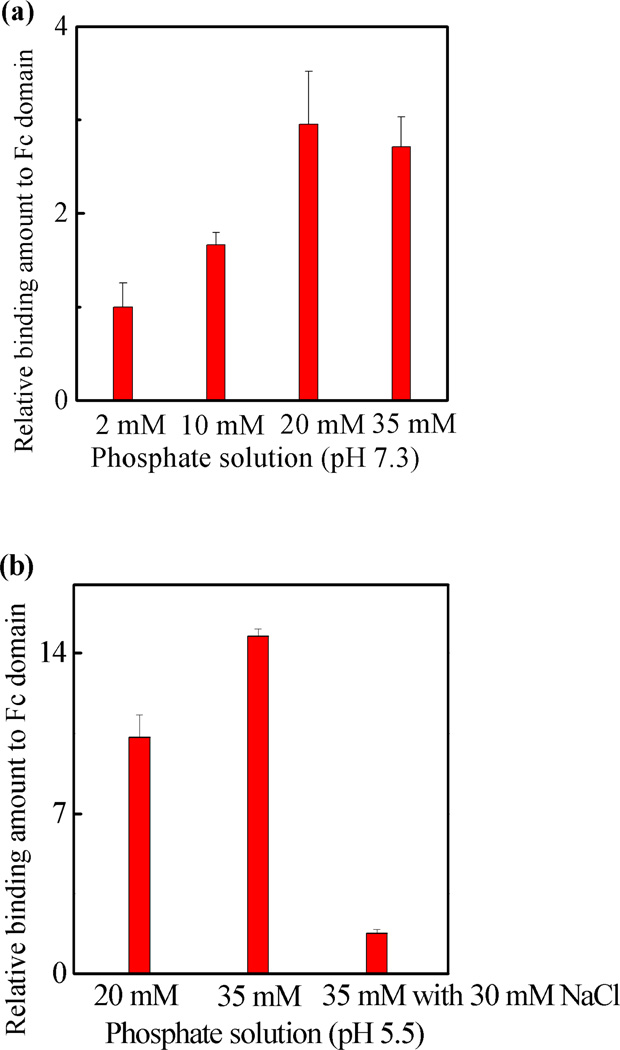
The preceding experiments provided evidence for a strong interaction between NP7 with the Fc domain of IgG. The composition of protein and functional polymer NP surfaces renders these interactions susceptible to binding conditions such as pH and ionic strength. The response to these variables can provide information about the nature of the binding.31–35 The influences of pH and ionic strength on the NP7-Fc interaction are summarized in Figure 5. In phosphate buffer (pH 7.3, without NaCl), an increase in phosphate concentration from 2 mM to 35 mM resulted in a three fold increase in the amount of protein bound to NP7 (Figure 5a, note that pH of the solution did not change even in the low buffer concentration). This implies that under the specified conditions the dominant contribution to binding is from hydrophobic interactions.32 A comparison of Figures 5a and b, reveals the binding between the Fc domain and NP7 is at least twice as strong at pH 5.5 than pH 7.3 in 20 mM phosphate buffer (without NaCl). At pH 7.3, exposed lysine (pKa 10.79) and arginine residues (pKa 12.48) are positively charged but at pH 5.5 and below histidine (pKa 6.04) is also positively charged. A mixture composed mainly of IgG1 and IgG2 are used in these studies. Assuming the five exposed histidine residues on the Fc domain are positively charged at or below pH 5.5, the human IgG1 Fc domain is estimated to have a net charge of +1 at neutral pH but +6 at or below pH 5.5 (see table S3). Similarly, the human IgG2 Fc domain has a net charge of 0 at neutral pH, but a net charge of +6 at or below pH 5.5. On the basis of simple electrostatic arguments, NP7 is expected to have a stronger interaction for the Fc domain at pH 5.5. Some support for this comes from the effect of ionic strength on binding. It was noted that the binding amount at pH 5.5 decreased as the NaCl salt concentration of 35 mM phosphate buffer was raised from 0 mM to 30 mM. This response is typical of a binding with contributions from electrostatic interactions.34,35
Figure 5.
The interaction between the Fc domain and NP7 by QCM at various phosphate concentrations at (a) pH 7.3; and (b) pH 5.5. The relative amount is related to the amount of NP7 bound to the Fc domain at various salt concentrations compared to the amount bound in 2 mM phosphate buffer at pH 7.3. Standard deviations were calculated from three independent measurements.
Inhibition of protein A-Fc binding by NP7
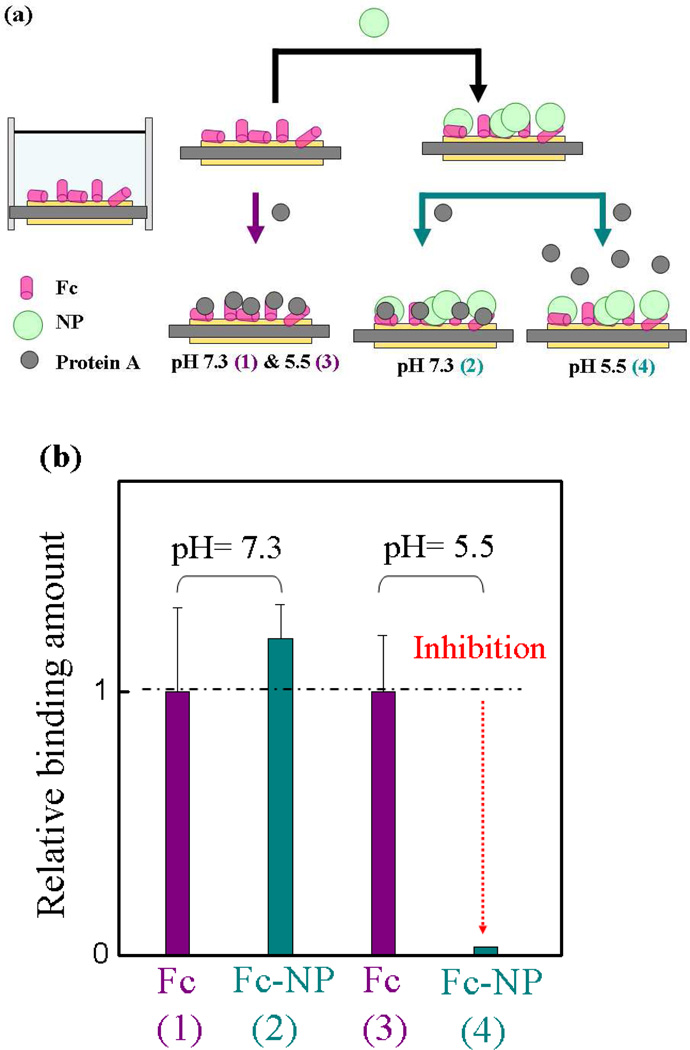
Protein A binds to the Fc domain of IgG. This strong and specific affinity is utilized for antibody purification. An X-ray crystal structure of the protein A-Fc complex identifies the protein-protein interaction site and the specific interactions that are responsible for binding.36 In the crystal, fragment B of protein A forms two important contacts with the Fc domain, one predominantly hydrophobic and the second is mainly polar. While our intention was not to directly mimic protein A-Fc binding, we anticipated that knowledge of the strength and location of this interaction would be helpful in analyzing the results of the NP binding studies (vide infra). To this end a competitive inhibition experiment between protein A and NP7 to the Fc domain of IgG under various buffer conditions was carried out. Figure 6 shows the experimental design and results of the competition at pH 7.3 and 5.5. The QCM cell surface was functionalized with Fc fragments. NPs were introduced into the cells at pH 5.5 and 7.3, respectively, until saturation of the signal was observed. Protein A was then injected into the cells and the response to the added protein was monitored. A control experiment was performed following the same procedure except the NPs were omitted. The results show that at pH 7.3, NP7 has little effect on protein A binding to the Fc domain. In contrast, at pH 5.5, preincubation of the Fc domain with NP7 inhibits protein A binding (~ 97% inhibition, Figure 6b column 4). Non-specific binding between NP7 and protein A at pH 7.3 and 5.5 were also considered. We attribute the difference in association between protein A and NP7 to the change in net charge of the NP/protein A binding domain of Fc due to the status of protonation of histidine residues (vide infra). Furthermore, the experimental results indicate that NP7 and protein A can compete for the same binding site at pH 5.5.
Figure 6.
(a) Schematic of the competitive binding of protein A to Fc in the presence and absence of NP7. (b) The interaction of the Fc fragment and protein A at pH 7.3 and 5.5 in PBS (35 mM, without salt), respectively, following treatment with NP7. The arrow color in (a) corresponds to the color of the column in (b). Cyan column: the frequency change resulting from protein A binding to the Fc domain in the presence of NP7 at pH 7.3 (2) and 5.5 (4), respectively. Purple column: control experiment under the same conditions omitting NPs at pH 7.3 (1) and 5.5 (3), respectively. Relative binding is defined by comparing the binding amount of the NP to that of protein A for the Fc domain. Standard deviations were calculated from three independent measurements.
NP7 Inhibition of protein A binding to IgG
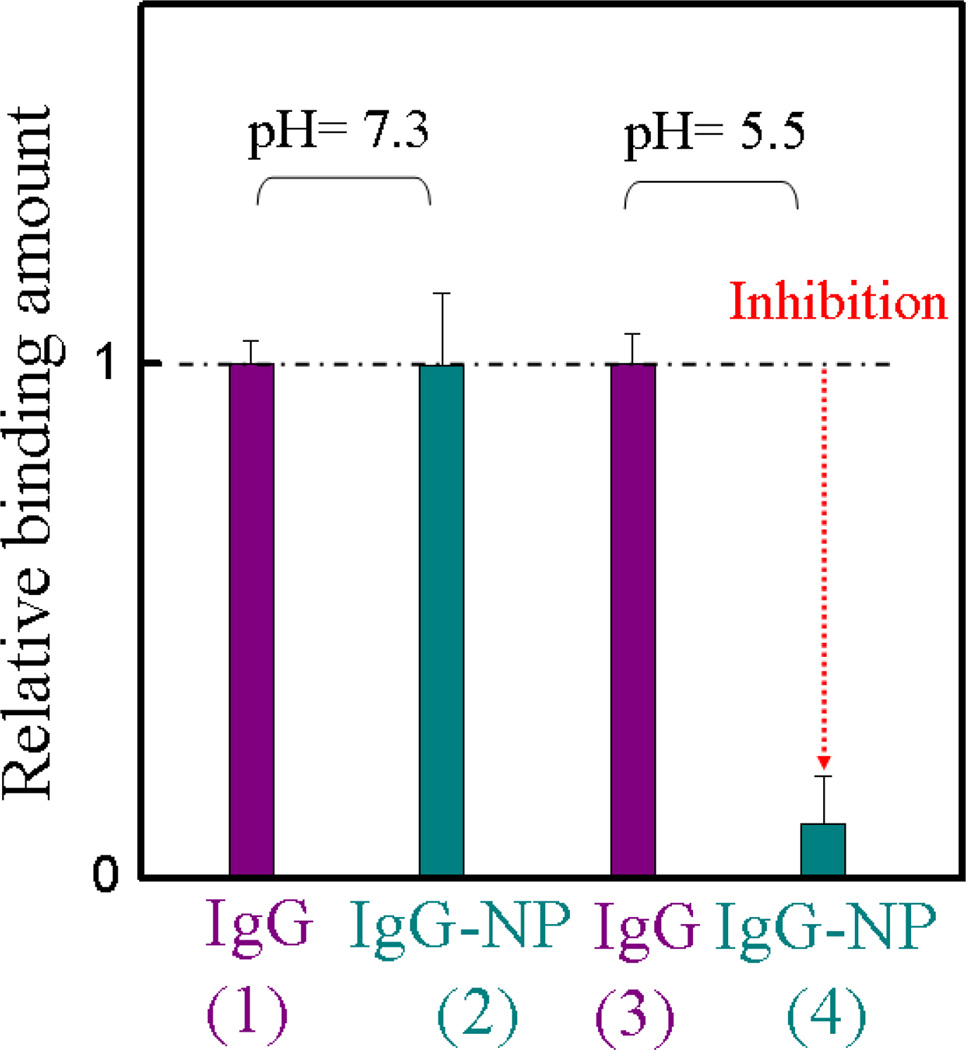
The preceding results suggest that NP7 competes with protein A at a discrete Fc domain at pH 5.5. To establish if this result is also observed with an intact IgG, a procedure similar to that described in Figure 6a was carried out only the Fc domain was replaced with IgG in the immobilization step (Experimental Methods). NP7 was injected into the cell with immobilized IgG until signal saturation was observed. A solution of protein A was subsequently injected. The control experiment consisted of the same procedure except the addition of NPs was omitted. At pH 7.3, there was no difference between cells with or without NP7. The results are quite similar to the previous experiment with the Fc fragment, no inhibition (competition) between NP7 with protein A at pH 7.3. In contrast, at pH 5.5, there is little interaction between protein A and Fc after IgG was treated with NP7 (Figure 7, cyan, column 4). We conclude that the NP inhibits protein A binding at pH 5.5. This phenomenon is analogous to the pH-dependent interaction of the neonatal Fc receptor (FcRn) to IgG.37–40 It exhibits high-affinity binding at pH 6.0–6.5 but weaker binding at pH 7.0–7.5. FcRn binds maternal IgG from ingested milk in the gut (pH 6.0–6.5) and delivers it to the bloodstream of the newborn (pH 7.0–7.5). In a process, called transcytosis, IgG proteins are transported across the gut epithelium to enter the bloodstream of the neonate. Many studies38–40 have shown that the pH response is due to histidine residues on the Fc domain. Both the FcRn and protein A binding sites are at the interface between the CH2–CH3 domains (vida infra) of IgG. It is also known that fragment B of protein A inhibits FcRn binding to IgG.41 This indicates there is overlap in the binding domains of FcRn and protein A to the Fc domain of IgG. In addition, research has shown that two histidines39,41 (Fc residues 310 and 433) are responsible for the pH dependent binding, and they are also located on fragment B of the protein A-IgG binding interface. The similar pH response of NP7 and FcRn binding to the Fc domain supports the hypothesis that the pH-dependent stems from interactions with His residues.
Figure 7.
Interaction between the Fc fragment of IgG and protein A at pH 7.3 and 5.5 in 35 mM phosphate buffer, before and after addition of NP7. The experimental design is similar to Figure 6a. Whole IgG was used to functionalize the QCM cell. The color of the column in the graph corresponds to the color of the arrowhead in Figure 6a. Cyan column: the frequency change was caused by protein A binding to the Fc domain of IgG in the presence of NP7 at pH 7.3 (2) and 5.5 (4), respectively. Purple column: control experiment under the same conditions except omitting the introduction of NPs at pH 7.3 (1) and 5.5 (3). Relative binding is defined as comparing the binding amount of the NP to that of protein A for whole IgG. Standard deviations were calculated from three independent measurements.
Computational study of the Fc surface and analysis of potential NP7 binding sites
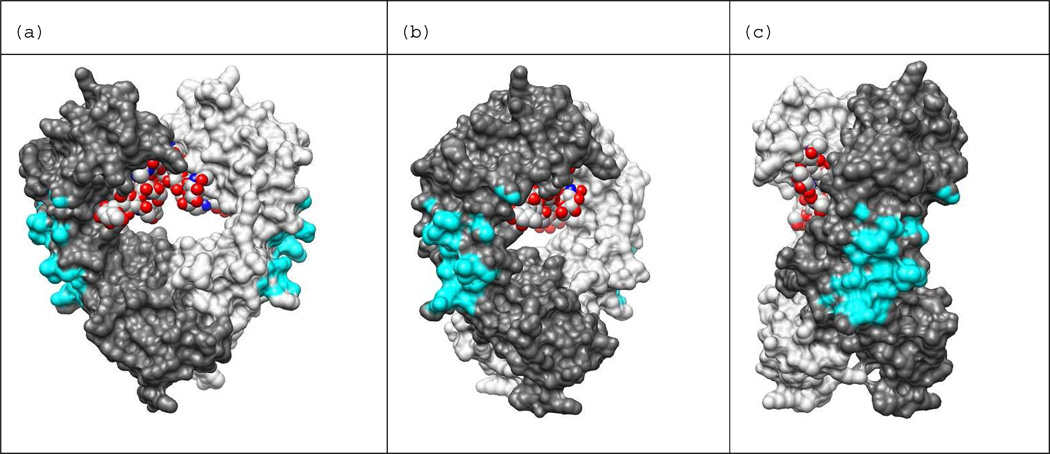
To understand the origins of the inhibition of protein A-Fc binding by NP7, the binding surface of the protein A-Fc complex was analyzed from the X-ray crystal structure of the complex. The analysis identifies the protein-protein interaction site and the specific residues that are responsible for binding.36 The interaction occurs in the hinge region between the CH2 and CH3 domains. Upon binding protein A, 660 Å2 of SASA on Fc is covered. Figure 8 presents various views of the Fc dimer with the atoms involved in binding protein A shown in cyan (an atom in Fc was defined as in contact with protein A if it was within 5 Å of any heavy atom of protein A). The experimental structure of protein A bound to Fc (PDB ID: 1FC2) was used to calculate the contacting atoms, and the images in Figure 8 were generated from the apo structure (PDB: 1HZH) using UCSF Chimera.42 Using the 5 Å distance for defining contact, 77 heavy atoms in Fc are in contact with protein A. The interface between protein A and Fc is stabilized mainly by hydrophobic interactions, with relatively few polar contacts.18 The hinge region between the CH2 and CH3 domains of Fc also contains ample polar groups allowing for high affinity binding to biochemically diverse surfaces.
Figure 8.
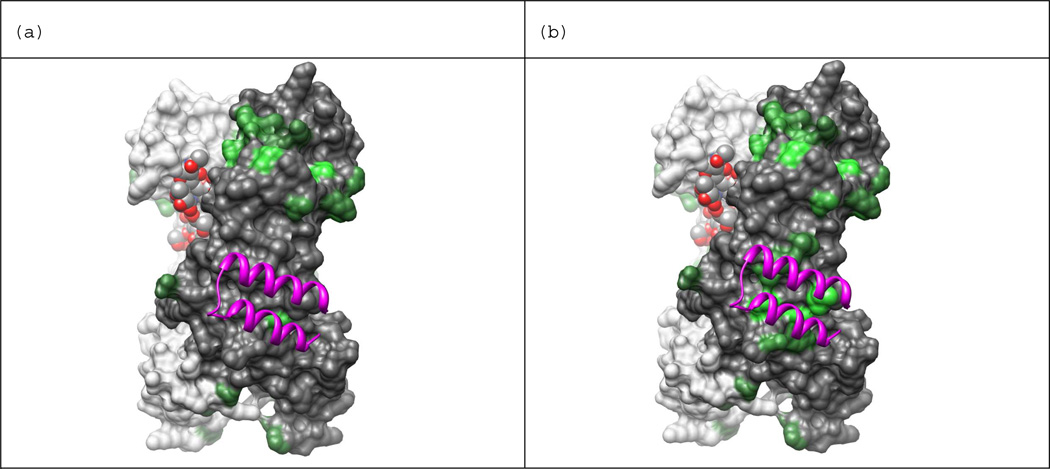
Protein A binding site shown on IgG1-Fc dimer. Binding site atoms are colored cyan. An Fc atom is considered in contact with protein A if it is within 5 Å of any heavy atom of protein A. The holo structure PDB ID: 1FC2 was used to calculate distances between Fc and protein A. The N-terminus of Fc is at the top of the view and C-terminus is at the bottom. Chain H is colored dark gray, chain K is colored light gray, and the carbohydrates bound in the Fc dimer core (red) are shown in the space filled representation. Images generated from PDB ID: 1HZH with UCSF Chimera.42 The view in (b) is rotated 45 degrees about the vertical axis with respect to the view in (a), and the view in (c) is rotate 90 degrees with respect to (a).
A computational study of the Fc surface was performed to search for likely NP7 binding regions to assist in interpreting the pH dependence of the protein A-NP competition study. In addition to hydrogen bond donor and acceptor groups, NP7 is composed of hydrophobic and negatively charged groups, thus, the Fc protein surface was scanned for complementary regions (hydrophobic and positively charged) as described in the Experimental Methods. The calculated charge and hydrophobicity scores for all Fc residues are presented in Table S4. To distill the surface region calculations to a single value, the combined score of a surface region was defined as: 2 × charge score + hydrophobic score. The combined score was set to 0 for surface regions with charge score • 0. The SASA, hydrophobic score, charge score, and combined score for all 97 surface regions are presented in Table S5.
Modeling of potential NP7 binding sites at neutral pH
The surface region with the highest combined score (+8.8) at neutral pH is centered on Thr289 in the CH2 domain of Fc. This region has SASA of 669 Å2, it includes 5 positively charged residues (Lys274, Lys288, Lys290, Lys292, and Lys301) and only one negatively charged residue (Glu272), giving it a charge score of +4. The hydrophobicity score is +0.8 and the partially exposed hydrophobic residues in the region include: Phe275, Val284, Ala287, Val303, and Val305. Figure 9 depicts a view of the Fc dimer, highlighting the centers of the surface regions with the highest combined scores. Figure 9a depicts all surface regions with a combined score > 0 at neutral pH in green, with increased brightness corresponding to higher values. Notice that, in addition to the brightest green surface (Thr289) many other residues on the CH2 domain are also green. It is likely that these residues in CH2 form a contiguous interaction interface with NP7. These results indicate that protein A (shown as purple ribbons) and NP7 are unlikely to compete for the same binding site at neutral pH. This is consistent with the results of the inhibition study.
Figure 9.
Computationally selected NP binding sites at (a) neutral and (b) low pH. Computational results indicate NP7 is more likely to bind the protein A binding site at lower pH in agreement with experimental observations. Residue side-chains are colored green if the combined score of its surface region is greater than 0.0, and brighter shades of green indicate higher scores. (a) presents results calculated assuming neutral pH and (b) at low pH where His residues can carry a positive charge. Chain H is colored dark gray, chain K is colored light gray, and the carbohydrates bound in the Fc dimer core are shown in space fill representation. Protein A appears as purple ribbons. Images generated from PDB ID: 1HZH with UCSF Chimera.42
Modeling of potential NP7 binding sites at low pH
The surface region with the highest combined score (+9.4) at low pH is centered on Met252 in the hinge region between the CH2 and CH3 domains of Fc. This region has SASA of 812 Å2, it includes 5 positively charged residues (Lys246, Lys248, Arg255, His310, and His435) and two negatively charged residue (Asp249 and Glu380), giving it a charge score of +3. The hydrophobicity score is +3.4 and exposed hydrophobic surface includes the fully exposed side-chain of Ile253 and the partially exposed side-chains of Met252 and Leu314. Figure 9b depicts all surface regions with a combined score > 0 at low pH in green with increased brightness corresponding to higher values. The surface region centered on Met252 appears in bright green in the area covered by protein A. In addition to Met252, several other residues in the protein A binding region also appear in green. It is likely that these residues in the hinge region form a contiguous interaction interface with NP7. These results suggest that protein A and NP7 are likely to compete for the same binding site at lower pH and are consistent with the data obtained from the binding studies.
Affinity of NP7 to the Fc domain
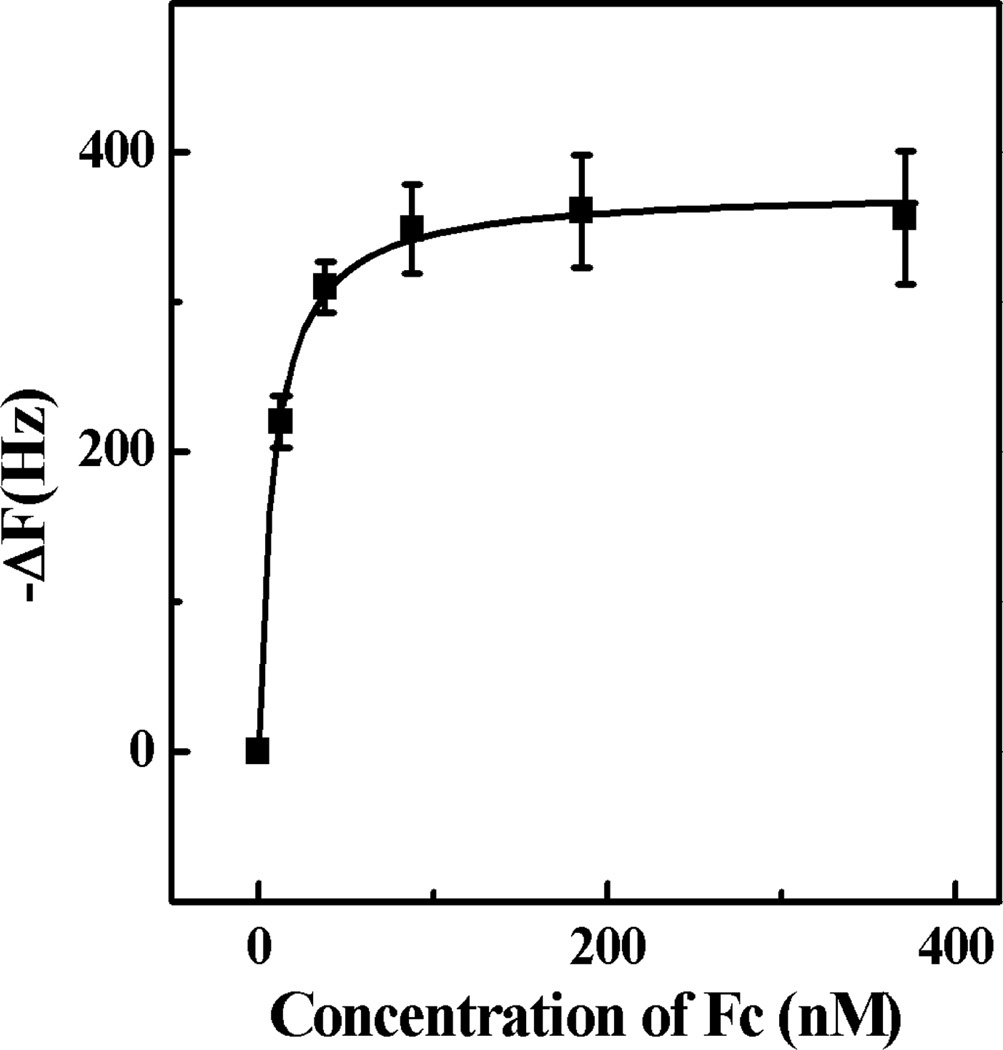
To calculate the binding affinity of IgG and NP7, IgG was immobilized on a QCM cell surface and NPs were injected in aliquots until signal saturation was achieved (Figure 10). A frequency change of approximately 3000 Hz was observed corresponding to almost complete coverage of the surface by NP7.43 Washing the cells resulted in a slight diminution of frequency (less than 2%). A solution of the Fc fragment was then added to the cells in aliquots and the resulting change in frequency was monitored. The data was fitted to a Langmuir isotherm from which a Kd of 8.5 nM for the NP-Fc fragment was obtained. A second method where the Fc fragment was immobilized was also used to obtain Kd (Figure S6). The molecular weight of NP7 obtained by static light scattering is 1.02 × 104 kDa (Figure S9). A calculated apparent dissociation constant of 3.5 nM was obtained by nonlinear fitting of the binding isotherm to a Langmuir isotherm. The two dissociation constants measured by these independent experiments are in good agreement. The dissociation constant of protein A is 10 nM17 at pH 7. These results independently indicate that NP7 has a comparable affinity to IgG at pH 5.5 as protein A at pH 7. These results suggest that synthetic polymers with a composition of NP7 have the potential to be used as an affinity media for IgG.
Figure 10.
Binding isotherm of Fc domain to NP7 immobilized on QCM surface in water (pH 5.5) Standard deviations were calculated from three independent measurements.
CONCLUSION
An iterative process was used to identify synthetic polymer NPs (50~65 nm) composed of 40% TBAm, 20% AAc, 2% Bis and 38% NIPAm that binds to the Fc fragment of IgG. The binding was found to be pH sensitive. The binding amount of the synthetic polymer NPs to Fc at pH 5.5 is twice that at pH 7.3 in 20 mM phosphate buffer. The difference in binding is attributed to the difference in the charged state of histidine residues of Fc. At pH 5.5, NP7 can inhibit protein A binding to the Fc domain. However, there was no such competition observed at pH 7.3. A computational analysis identified potential binding domains. At pH 5.5, a binding domain of NP7 overlaps with the protein A-Fc binding domain. In addition, the affinity of NP7 at pH 5.5 is comparable to protein A at pH 7. We have also presented preliminary evidence that NP7 can capture the whole IgG. At pH 5.5, NP7 competes with the binding of protein A to IgG in a similar fashion to that of the Fc fragment. These results suggest that synthetic polymer NPs can be engineered to have an intrinsic affinity to a specific domain of a large biomacromolecule.
Supplementary Material
ACKNOWLEDGMENT
We are grateful for financial support from NIH (NIH GM080506), The Shiseido Corp, MEXT (23111716) and JSPS (23750193). We also thank Initium, Inc. for QCM support. SHL was supported by a National Science Council Taiwan graduate student study abroad fellowship. AR was supported by a NIH/NLM Pathway to Independence Award (K99LM010821); PB was supported by NIH grants LM010235-01A1 and 5T15LM007743. We also wish to thank Mr. Isao Yanagisawa (Shiseido).
ABBREVIATIONS
- NP
nanoparticle
- Bis
N,N’-methylene-bisacrylamide
- AAc
acrylic acid
- APM
N- (3-aminopropyl) methacrylamide hydrochloride
- NIPAm
N-isopropylacrylamide
- ECD
N- (3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride
- BSA
bovine serum albumin
- TNFα
tumor necrosis factor-α human
- SDS
sodium dodecyl sulfate
- CTAB
cetyltrimethylammonium bromide
- AIBN
azobisisobutyronitrile
- NIH
N-Hydroxysuccinimide
- QCM
Quartz crystal microbalance
- DLS
dynamic light scattering
- NIH
National Institutes of Health
Footnotes
ASSOCIATED CONTENT
SUPPORTING INFORMATION
Supporting Information Available: Experimental methods for NPs preparation, confirmation of Fc-orientation immobilization, model calculation, competitive binding of protein A to Fc, binding isotherm of NP 7 to the Fc. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Meenach SA, Hilt JZ, Anderson KW. Acta Biomater. 2010;6:1039–1046. doi: 10.1016/j.actbio.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Bhojani MS, Van Dort M, Rehemtulla A, Ross BD. Mol. Pharmaceut. 2010;7:1921–1929. doi: 10.1021/mp100298r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tong R, Christian DA, Tang L, Cabral H, Baker JR, Kataoka K, Discher DE, Cheng JJ. Mrs Bull. 2009;34:422–431. [Google Scholar]
- 4.Messerschmidt SKE, Musyanovych A, Altvater M, Scheurich P, Pfizenmaier K, Landfester K, Kontermann RE. J. Control. Release. 2009;137:69–77. doi: 10.1016/j.jconrel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 6.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Angew. Chem., Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 7.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat. Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 8.Horgan AM, Moore JD, Noble JE, Worsley GJ. Trends Biotechnol. 2010;28:485–494. doi: 10.1016/j.tibtech.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino Y, Urakami T, Kodama T, Koide H, Oku N, Okahata Y, Shea KJ. Small. 2009;5:1562–1568. doi: 10.1002/smll.200900186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, Shea KJ. J. Am. Chem. Soc. 2010;132:6644–6645. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino Y, Koide H, Furuya K, Haberaecker WW, Lee SH, Kodama T, Kanazawa H, Oku N, Shea KJ. PNAS. 2012;109:33–38. doi: 10.1073/pnas.1112828109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You CC, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimatsu K, Lesel BK, Yonamine Y, Beierle JM, Hoshino Y, Shea KJ. Angew. Chem. Int. Ed. 2012;51:2455–2458. doi: 10.1002/anie.201107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. https://commonfund.nih.gov/proteincapture/
- 15.Roque ACA, Silva CSO, Taipa MA. J. Chromatogr. A. 2007;1160:44–55. doi: 10.1016/j.chroma.2007.05.109. [DOI] [PubMed] [Google Scholar]
- 16.Miyakawa S, Nomura Y, Sakamoto T, Yamaguchi Y, Kato K, Yamazaki S, Nakamura Y. RNA. 2008;14:1154–1163. doi: 10.1261/rna.1005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 18.Sauereriksson AE, Kleywegt GJ, Uhlén M, Jones TA. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 19.(a) Chee CK, Rimmer S, Soutar I, Swanson L. Polymer. 2001;42:5079–5087. [Google Scholar]; (b) Ottaviani MF, Winnik FM, Bossmann SH, Turro NJ. Helv. Chim. Acta. 2001;84:2476–2492. [Google Scholar]
- 20.Hoshino Y, Haberaecker WW, Kodama T, Zeng ZY, Okahata Y, Shea KJ. J. Am. Chem. Soc. 2010;132:13648–13650. doi: 10.1021/ja1058982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roque ACA, Lowe CR, Taipa MA. Biotechnol. Prog. 2004;20:639–654. doi: 10.1021/bp030070k. [DOI] [PubMed] [Google Scholar]
- 22.Low D, O'Leary R, Pujar NS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;848:48–63. doi: 10.1016/j.jchromb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Thommes J, Etzel M. Biotechnol. Prog. 2007;23:42–45. doi: 10.1021/bp0603661. [DOI] [PubMed] [Google Scholar]
- 24.Nematollahzadeh A, Sun W, Aureliano CSA, Lutkemeyer D, Stute J, Abdekhodaie MJ, Shojaei A, Sellergren B. Angew. Chem. Int. Ed. 2011;50:495–498. doi: 10.1002/anie.201004774. [DOI] [PubMed] [Google Scholar]
- 25.Ren LB, Liu Z, Liu YC, Dou P, Chen HY. Angew. Chem., Int. Ed. 2009;48:6704–6707. doi: 10.1002/anie.200902469. [DOI] [PubMed] [Google Scholar]
- 26.Debord JD, Lyon LA. Langmuir. 2003;19:7662–7664. [Google Scholar]
- 27.Ogawa K, Nakayama A, Kokufuta E. Langmuir. 2003;19:3178–3184. [Google Scholar]
- 28.Song D, Forciniti D. J. Colloid Interface Sci. 2000;221:25–37. doi: 10.1006/jcis.1999.6560. [DOI] [PubMed] [Google Scholar]
- 29.Jans H, Liu X, Austin L, Maes G, Huo Q. Anal. Chem. 2009;81:9425–9432. doi: 10.1021/ac901822w. [DOI] [PubMed] [Google Scholar]
- 30.Peppel K, Crawford D, Beutler B. J. Exp. Med. 1991;174:1483–1489. doi: 10.1084/jem.174.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goncalves E, Kitas E, Seelig J. Biochemistry. 2006;45:3086–3094. doi: 10.1021/bi052221t. [DOI] [PubMed] [Google Scholar]
- 32.Lin FY, Chen CS, Chen WY, Yamamoto S. J. Chromatogr. A. 2001;912:281–289. doi: 10.1016/s0021-9673(01)00584-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang SP, Sun Y. Biotechnol. Bioeng. 2001;75:710–717. doi: 10.1002/bit.10067. [DOI] [PubMed] [Google Scholar]
- 34.De M, You CC, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 35.Zeng ZY, Patel J, Lee SH, McCallum M, Tyagi A, Yan MD, Shea KJ. J. Am. Chem. Soc. 2012;134:2681–2690. doi: 10.1021/ja209959t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deisenhofer J. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 37.Raghavan M, Bjorkman PJ. Annu. Rev. Cell Dev. Biol. 1996;12:181–220. doi: 10.1146/annurev.cellbio.12.1.181. [DOI] [PubMed] [Google Scholar]
- 38.Kim JK, Tsen MF, Ghetie V, Ward ES. Eur. J. Immunol. 1994;24:2429–2434. doi: 10.1002/eji.1830241025. [DOI] [PubMed] [Google Scholar]
- 39.Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Biochemistry. 1995;34:14649–14657. doi: 10.1021/bi00045a005. [DOI] [PubMed] [Google Scholar]
- 40.Martin WL, West AP, Gan L, Bjorkman PJ. Mol. Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 41.Raghavan M, Chen MY, Gastinel LN, Bjorkman PJ. Immunity. 1994;1:303–315. doi: 10.1016/1074-7613(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Ozeki T, Morita M, Yoshimine H, Furusawa H, Okahata Y. Anal. Chem. 2007;79:79–88. doi: 10.1021/ac060873x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.