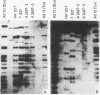
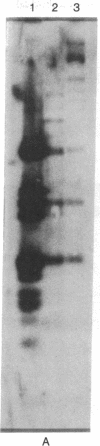
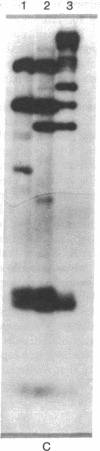
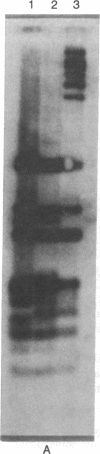
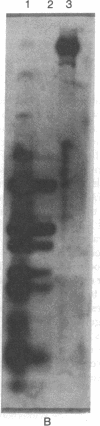
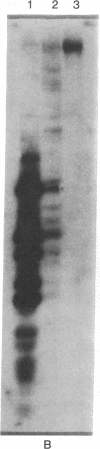
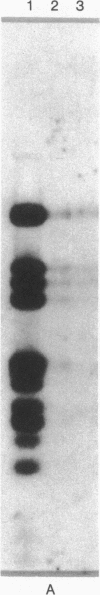
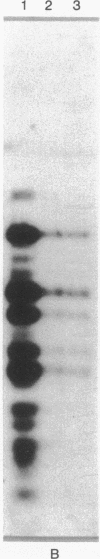
Abstract
The adenovirus type 12 (Ad12) DNA sequences integrated into the DNA of four lines of Ad12-transformed hamster cells are extensively methylated. Methylation in mammalian cell DNA is believed to occur predominantly at 5'-C-G-3' sequences. The majority, although not all, of the 5'-C-C-G-G-3' sequences present in integrated Ad12 DNA are methylated. Ad12 DNA isolated from purified virions, on the other hand, is not methylated to any significant extent. The segments of the integrated viral DNA comprising early genes, which are expressed as mRNA in two lines of Ad2-transformed hamster cells, are undermethylated in comparison to late viral segments, which are not expressed and are extensively methylated. In contrast, in two lines of Ad12-induced rat brain tumor cells, some of the late viral genes have been shown to be expressed as mRNA. The segment of the integrated Ad12 DNA that comprises these late genes, the EcoRI B fragment, is undermethylated in comparison to the extensive methylation of the same fragment in Ad12-transformed hamster cells. Thus, there appears to exist a striking inverse correlation between the levels of methylation of specific DNA segments and the extent to which these segments are expressed as mRNA. The functional significance of this correlation remains to be determined. It may provide a clue to understanding the regulation of gene expression in transformed cells and perhaps in eukaryotic cells in general.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W., Lundholm U., Hirsch-Kauffmann M. Intracellular forms of adenovirus deoxyribonucleic acid. I. Evidence for a deoxyribonucleic acid-protein complex in baby hamster kidney cells infected with adenovirus type 12. J Virol. 1972 Feb;9(2):297–308. doi: 10.1128/jvi.9.2.297-308.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Gunthert U., Schweiger M., Stupp M., Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Ortin J., Scheidtmann K. H., Greenberg R., Westphal M., Doerfler W. Transcription of the genome of adenovirus type 12. III. Maps of stable RNA from productively infected human cells and abortively infected and transformed hamster cells. J Virol. 1976 Nov;20(2):355–372. doi: 10.1128/jvi.20.2.355-372.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Scheidtmann K. H., Mansi-Wothke H., Doerfler W. Transcription of the genome of adenovirus type 12. V. Kinetic and size-class analyses of nuclear and messenger RNA in productive infection. J Gen Virol. 1979 Sep;44(3):699–713. doi: 10.1099/0022-1317-44-3-699. [DOI] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- WYATT G. R. The purine and pyrimidine composition of deoxypentose nucleic acids. Biochem J. 1951 May;48(5):584–590. doi: 10.1042/bj0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]