ABSTRACT
Kingella kingae is an emerging bacterial pathogen that is being recognized increasingly as an important etiology of septic arthritis, osteomyelitis, and bacteremia, especially in young children. Colonization of the posterior pharynx is a key step in the pathogenesis of K. kingae disease. Previous work established that type IV pili are necessary for K. kingae adherence to the respiratory epithelium. In this study, we set out to identify additional factors that influence K. kingae interactions with human epithelial cells. We found that genetic disruption of the gene encoding a predicted trimeric autotransporter protein called Knh (Kingella NhhA homolog) resulted in reduced adherence to human epithelial cells. In addition, we established that K. kingae elaborates a surface-associated polysaccharide capsule that requires a predicted ABC-type transporter export operon called ctrABCD for surface presentation. Furthermore, we discovered that the presence of a surface capsule interferes with Knh-mediated adherence to human epithelial cells by nonpiliated organisms and that maximal adherence in the presence of a capsule requires the predicted type IV pilus retraction machinery, PilT/PilU. On the basis of the data presented here, we propose a novel adherence mechanism that allows K. kingae to adhere efficiently to human epithelial cells while remaining encapsulated and more resistant to immune clearance.
IMPORTANCE
Kingella kingae is a Gram-negative bacterium that is being recognized increasingly as a cause of joint and bone infections in young children. The pathogenesis of disease due to K. kingae begins with bacterial colonization of the upper respiratory tract, and previous work established that surface hair-like fibers called type IV pili are necessary for K. kingae adherence to respiratory epithelial cells. In this study, we set out to identify additional factors that influence K. kingae interactions with respiratory epithelial cells. We discovered a novel surface protein called Knh that mediates K. kingae adherence and found that a surface-associated carbohydrate capsule interferes with the Knh-mediated adherence of organisms lacking pili. Further analysis revealed that pilus retraction is necessary for maximal Knh-mediated adherence in the presence of the capsule. Our results may lead to new strategies to prevent disease due to K. kingae and potentially other pathogenic bacteria.
Introduction
Kingella kingae is a Gram-negative bacterium in the family Neisseriaceae and was originally considered a rare cause of human disease. However, improvements in culture-based and molecular-analysis-based diagnostics over the past 2 decades have led to increased recognition of this organism as an emerging pathogen (1–8). K. kingae is the causative agent of a number of pediatric diseases, including septic arthritis, osteomyelitis, bacteremia, and endocarditis (6, 8). A recent report indicates that K. kingae is the leading cause of osteoarticular infections in children 6 to 36 months of age (1).
K. kingae initiates infection by colonizing the posterior pharynx and is a common commensal organism in young children (9, 10). The organism then enters the bloodstream and disseminates to distant sites, including joints, bones, and the endocardium (8, 11, 12). A key step in colonization of the respiratory tract is adherence to the respiratory epithelium. Previous studies have demonstrated that K. kingae type IV pili are essential for adherence to human epithelial cells (13). In particular, genetic disruption of the gene encoding the major pilin subunit called PilA1 results in loss of piliation and complete loss of adherence (13, 14).
Expression of type IV pili is phase variable, allowing for evasion of the immune system. Our prototypic parent K. kingae strain, 269-492, grows on solid agar as two phenotypically stable colony types, referred to as nonspreading/noncorroding colonies and spreading/corroding colonies. The nonspreading/noncorroding colony variant (designated KK01) expresses low levels of type IV pili, while the spreading/corroding colony variant (designated KK03) expresses high levels of type IV pili. In a collection of clinical isolates from diverse anatomic sites, we have also identified a domed colony type associated with no pili. Further analysis of this collection revealed that the majority of respiratory and nonendocarditis blood isolates were piliated, while the majority of joint fluid, bone, and endocarditis blood isolates were nonpiliated, suggesting a role for pilus phase variation in vivo (15).
While it is clear that type IV pili are important for adherence to human epithelial cells, the fact that a significant percentage of pharyngeal isolates are nonpiliated suggests that K. kingae expresses additional surface factors that modulate interactions with host cells. Accordingly, to gain a more thorough understanding of the bacterial factors that potentially influence colonization of the pharynx, we set out to identify additional determinants of adherence to human epithelial cells. This search revealed a novel trimeric autotransporter called Knh (Kingella NhhA homolog), which is necessary for full-level K. kingae adherence to human epithelial cells. Additional studies demonstrated that K. kingae elaborates a surface-associated polysaccharide capsule that interferes with Knh-mediated adherence. Analysis of a K. kingae mutant incapable of pilus retraction suggested that capsule-mediated interference is overcome by the retraction of type IV pili, presumably displacing polysaccharide and facilitating access of Knh to the host cell surface. This adherence mechanism allows K. kingae to adhere efficiently to host cells while remaining encapsulated and resistant to host immune clearance strategies.
RESULTS
K. kingae encodes a novel trimeric autotransporter.
K. kingae type IV pili are essential for adherence to human epithelial cells (13). To identify other factors that might contribute to K. kingae adherence, we examined the draft genome sequence of strain 269-492 for putative adhesive factors by searching for homologs of genes encoding adhesins in other Neisseriaceae family members, including the Neisseria meningitidis Opa, Opc, NadA, and NhhA proteins. This analysis revealed a 5,352-bp open reading frame (ORF) encoding a protein that has C-terminal sequence homology to the N. meningitidis NhhA trimeric autotransporter (accession no. AAK09243.1) and was named Knh (16, 17). Trimeric autotransporters are a family of adhesive outer membrane proteins classified largely on the basis of the amino acid sequence in the conserved C-terminal beta-barrel membrane anchor domain. Sequence alignment revealed strong membrane anchor domain homology between Knh and NhhA, as well as the Haemophilus influenzae Hia protein (accession no. AAC43721.2) (Fig. 1A). Additional analysis with SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) revealed a predicted N-terminal signal peptide corresponding to amino acids 1 to 54 and a predicted signal peptidase cleavage site between amino acids 54 and 55, consistent with the possibility that Knh is secreted and surface localized. Domain annotation of Knh was accomplished with the Domain Annotation of Trimeric Autotransporters program (http://toolkit.tuebingen.mpg.de/dataa) and demonstrated an ISneck2 domain with architecture similar to that of the ISneck1 domain that has been associated with adhesive activity in the H. influenzae Hia and Hsf proteins (18), a series of 19 YadA-like head domains predicted to form a beta-roll on the basis of the Yersinia enterocolitica YadA head domain crystal structure (19, 20), and a series of eight Trp ring domains spaced between the YadA-like head domain region and the membrane anchor (Fig. 1B).
FIG 1 .
K. kingae Knh is a trimeric autotransporter. (A) Knh C-terminal membrane anchor amino acid sequence alignment with N. meningitidis NhhA and H. influenzae Hia. Asterisks indicate identical residues, colons indicate conserved substitutions, and periods indicate semi-conserved substitutions. (B) Annotation of Knh, highlighting the presence of numerous protein domains, including a signal peptide, a membrane anchor, YadA-like head domains, Trp ring domains, and an ISneck2 domain. (C) Strep-tag Western blot analysis of outer membrane preparations and formic acid-denatured outer membrane preparations following the induction of vector control pASK-IBA12 or pASK::Knh1695-1783, which expresses the Knh beta-barrel domain fused to a Strep-tag. Molecular sizes (MW) are expressed in kilodaltons.
To confirm that Knh is a trimeric autotransporter, the ability of the C-terminal beta-barrel domain to trimerize in the outer membrane was examined. A 264-nucleotide fragment of knh encoding amino acids 1695 to 1783, corresponding to the predicted beta-barrel membrane anchor, was cloned in frame into pASK-IBA12 (IBA BioTAGnology), which contains an N-terminal Escherichia coli OmpA signal sequence fused to a Strep-tag. The resulting construct, designated pASK::Knh1695-1783, was transformed into E. coli BL21 omp8 for induction. After secretion through the inner membrane and cleavage of the OmpA signal sequence, the recombinant Strep-tagged Knh1695-1783 protein has a predicted monomeric molecular mass of 12 kDa. As is shown in Fig. 1C, induction of pASK::Knh1695-1783 resulted in the outer membrane localization of a 36-kDa band detected with an anti-Strep monoclonal antibody. As has previously been reported with trimeric autotransporter proteins (21), the trimer was extremely stable under standard boiling and SDS-PAGE denaturing conditions and required an additional formic acid denaturation step to dissociate into monomers (Fig. 1C). These findings establish that the Knh beta-barrel is sufficient for trimerization in the outer membrane and confirm that Knh is a member of the growing class of trimeric autotransporter proteins.
Knh is expressed in the K. kingae outer membrane and is required for full-level adherence to human epithelial cells.
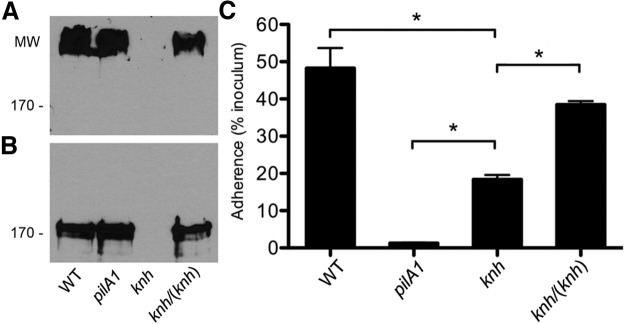
To determine if Knh is expressed and present in the outer membrane of K. kingae strain KK03, sarkosyl-insoluble membrane fractions were isolated and examined by Western blotting with anti-Knh serum. Strain KK03 is a naturally occurring derivative of our prototype K. kingae strain, 269-492, which expresses stable levels of type IV pili and was used as the wild-type (WT) strain throughout this study. As a negative control, a Knh mutant was created by insertionally inactivating knh in strain KK03. Under standard SDS-PAGE denaturing conditions, a high-molecular-mass band was evident in outer membrane preparations of strain KK03 and the nonpiliated, nonadherent pilA1 mutant but not in those of the knh mutant (Fig. 2A). While the high molecular mass of this reactive band precludes accurate size estimation, it is consistent with the trimer form of Knh, which has a predicted molecular mass of 540 kDa. Formic acid treatment of outer membranes resulted in elimination of the high-molecular-mass band and detection of a band corresponding to the predicted monomeric molecular mass (180 kDa) of Knh in strains KK03 and KK03pilA1 but not in strain KK03knh (Fig. 2B). To complement the knh mutation, the knh ORF with the native promoter region was recombined into a separate locus in the genome in the KK03knh strain background (see Fig. S1 in the supplemental material for the complementation strategy used). (To date, plasmids that replicate in K. kingae are unavailable.) As shown in Fig. 2A and B, outer membrane localization of Knh was restored in complemented strain KK03knh/(knh). Semiquantitative extracellular pilus preparation analysis using strain KK03pilA1 as a negative control revealed no difference in piliation levels among strains KK03, KK03knh, and KK03knh/(knh) (data not shown).
FIG 2 .
Knh is an outer membrane protein required for full-level adherence to Chang human epithelial cells. (A) Western blot analysis of outer membrane preparations from K. kingae strains KK03 (WT), KK03pilA1, KK03knh, and KK03knh/(knh) under standard SDS-PAGE denaturing conditions. (B) Western blot analysis of formic acid-treated outer membrane preparations from K. kingae strains KK03 (WT), KK03pilA1, KK03knh, and KK03knh/(knh) under standard SDS-PAGE denaturing conditions. Molecular sizes (MW) are expressed in kilodaltons. (C) Adherence of K. kingae strains KK03 (WT), KK03pilA1, KK03knh, and KK03knh/(knh) to Chang human epithelial cells. Adherence assays were conducted in triplicate. Error bars indicate standard errors of the means. Statistical analyses were performed by using the unpaired t test to compare the adherence levels of two strains as follows: P < 0.05 (*) for KK03 versus KK03knh, KK03pilA1 versus KK03knh, and KK03knh versus KK03knh/(knh).
To determine if Knh is involved in mediating K. kingae adherence to host cells, we examined the adherence of strains KK03, KK03pilA1, KK03knh, and KK03knh/(knh) in assays with Chang human epithelial cells. As shown in Fig. 2C, elimination of Knh resulted in a significant reduction in the level of adherence, compared to that of strain KK03 (P < 0.05), and complementation of the knh mutation almost completely restored adherence. Unlike the situation when type IV pili were eliminated by disruption of the pilA1 gene encoding the major pilin subunit, the disruption of knh did not completely abrogate adherence (P < 0.05 for KK03pilA1 versus KK03knh), suggesting that K. kingae type IV pili mediate low-level adherence and require Knh for full-level adherence.
Considered together, these data demonstrate that Knh is required for full-level adherence of K. kingae to Chang human epithelial cells and that Knh cannot mediate appreciable adherence in the absence of type IV pili.
K. kingae produces a surface-associated polysaccharide capsule.
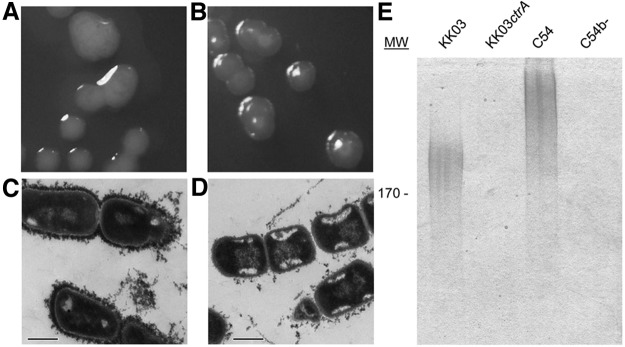
The observation that type IV pili are required for Knh-mediated adherence suggested that K. kingae may express another surface factor that blocks Knh adhesive activity. Examination of K. kingae strain KK03 revealed a mucoid colony phenotype similar to that seen with other bacterial species that produce a polysaccharide capsule (Fig. 3A). To explore the possibility that K. kingae expresses a polysaccharide capsule, the draft genome sequence was searched for homologs to genes involved in capsule synthesis and export in N. meningitidis, a well-studied Neisseriaceae family member that contains the siaABCD, ctrABCD, ctrE/lipA, and ctrF/lipB capsule-associated genes. As summarized in Table S1 in the supplemental material, an intact locus with homology to the ABC-type capsule export operon ctrABCD of N. meningitidis and unlinked genes with homology to the ctrE/lipA and ctrF/lipB genes were identified in the K. kingae genome. To determine if the ctrABCD operon plays a role in K. kingae encapsulation, ctrA was insertionally inactivated. As shown in Fig. 3B, examination of KK03ctrA revealed nonmucoid colonies, in marked contrast to the colonies of parental strain KK03. To confirm that K. kingae elaborates a polysaccharide capsule, strains KK03 and KK03ctrA were stained with cationic ferritin and then examined by thin-section transmission electron microscopy (TEM). Because of the highly anionic nature of most bacterial polysaccharide capsules, cationic ferritin typically binds to the capsular material, producing an electron-dense rim on the bacterial surface when visualized by thin-section TEM. A thick layer of electron density was evident on the surface of WT strain KK03 (Fig. 3C) and was absent from mutant strain KK03ctrA (Fig. 3D). Attempts to complement the mutation in ctrA and the likely polar effect on the ctrBCD genes were unsuccessful. Sequencing of our chromosomal complementation construct containing ctrABCD generated in E. coli consistently revealed nonsense or frameshift mutations in ctrA, presumably reflecting the inability of E. coli to tolerate the expression of the outer membrane protein encoded by ctrA. However, quantitative reverse transcription (qRT)-PCR analysis revealed that transcription of the ORF downstream of the ctrABCD operon was unaffected by the ctrA insertion mutation (see Fig. S2 in the supplemental material), supporting the conclusion that the phenotypes observed as a result of the ctrA mutation are due to disruption of the ctrABCD operon.
FIG 3 .
K. kingae has a surface capsule. (A, B) Colony phenotypes of K. kingae strains KK03 (A) and KK03ctrA (B). (C, D) Thin-section TEM analyses of cationic ferritin-stained K. kingae strains KK03 (C) and KK03ctrA (D). Scale bars, 500 nm. (E) Capsular material was extracted from strains KK03 and KK03ctrA with heat, separated by 7.5% SDS-PAGE, and then stained with the cationic dye alcian blue. Encapsulated H. influenzae type b strain C54 and an isogenic nonencapsulated mutant, C54b-, were included as controls. Molecular size (MW) is expressed in kilodaltons.
As a complementary approach to demonstrate the presence of a capsule, surface material was released from bacterial suspensions by heat extraction at 55°C, resolved by SDS-PAGE, and then stained with alcian blue (22). As shown in Fig. 3E, high-molecular-mass, alcian blue-reactive material was present in strain KK03 but absent from strain KK03ctrA. H. influenzae strain C54, which expresses a type b polysaccharide capsule, and H. influenzae strain C54b-, an isogenic mutant that is nonencapsulated, were used as controls. The alcian blue-reactive material did not stain with Coomassie blue and was unaffected by proteinase K treatment (data not shown), confirming that the extracted material is not proteinaceous.
Together, these data indicate that K. kingae strain KK03 expresses a surface-associated polysaccharide capsule that requires the predicted ctrABCD capsule export operon for surface localization.
Type IV pili, capsule, and Knh influence K. kingae adherence to human epithelial cells.
To further explore the interrelationship between type IV pili, Knh, and the capsule in terms of their effects on bacterial adherence, we generated single, double, and triple mutations in pilA1, knh, and ctrA. Extracellular pilus preparations revealed no change in pilus expression levels in mutant strains with an intact pilA1 gene (data not shown). As shown in Fig. 4, insertional inactivation of ctrA in strain KK03 resulted in a slight decrease in adherence that was not statistically significant. The adherence of the KK03ctrA/knh double mutant mimicked the levels seen in the knh single mutant, indicating that encapsulation does not influence type IV pilus-mediated adherence in the absence of Knh. Interestingly, while the pilA1 mutant was nonadherent, simultaneous disruption of pilA1 and ctrA in the KK03 background restored high-level adherence (P < 0.05 for the pilA1/ctrA mutant versus the pilA1 mutant), indicating that encapsulation inhibits pilus-independent Knh-mediated adherence. The pilA1/ctrA/knh triple mutant was nonadherent, providing strong evidence that Knh is an adhesin and is responsible for the K. kingae pilus-independent adherence observed in the pilA1/ctrA double mutant (P < 0.05 for the pilA1/ctrA/knh mutant versus the pilA1/ctrA mutant).
FIG 4 .
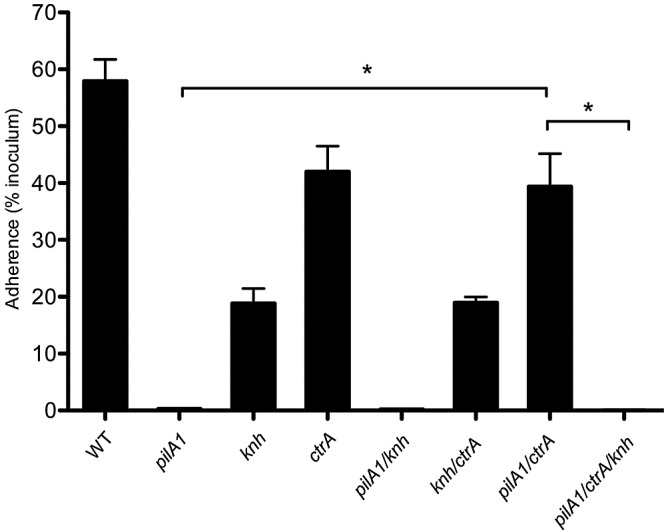
Influences of type IV pili, the capsule, and Knh on K. kingae adherence. The adherence of K. kingae KK03 (WT) and the pilA1, knh, ctrA, pilA1/knh, knh/ctrA, pilA1/ctrA, and pilA1/ctrA/knh mutant strains to Chang human epithelial cells was measured. Adherence assays were conducted in triplicate. Error bars indicate standard errors of the means. Statistical analyses were performed by using the unpaired t test to compare the adherence levels of two strains as follows: P < 0.05 (*) for pilA1/ctrA versus pilA1 and for pilA1/ctrA/knh versus pilA1/ctrA.
The type IV pilus retraction ATPase system is required for full-level adherence.
The observation that the capsule interferes with Knh-mediated adherence in the absence of pili but not in the presence of pili raised the possibility that pilus-mediated adherence draws the organism closer to the host cell surface, perhaps via pilus retraction. In accordance with this possibility, K. kingae encodes a homolog of the PilT/PilU retraction ATPase machinery that has been shown to be essential for pilus retraction in several well-studied type IV pilus-expressing bacteria, including N. meningitidis, N. gonorrhoeae, Pseudomonas aeruginosa, and Myxococcus xanthus (23–26). To determine if pilus retraction is necessary for full-level K. kingae adherence, pilT was insertionally inactivated in strain KK03. As the insertion in pilT likely had a polar effect on pilU, the pilTU operon with the native promoter was recombined into the chromosome to achieve complementation. Examination of KK03pilT revealed increased levels of extracellular pili, as assessed by semiquantitative pilus preparations (Fig. 5A), but lacked twitching motility, as assessed by a modification of the P. aeruginosa agar stab assay (Fig. 5B) (27), consistent with the hypothesis that the PilT/PilU retraction ATPase machinery is necessary for K. kingae pilus retraction. As shown in Fig. 5C, elimination of PilT/PilU resulted in adherence that was decreased compared to that of parent strain KK03 (P < 0.05) but equal to that of the knh mutant. Complementation of the pilTU operon in the pilT mutant background resulted in reduction of extracellular pilus expression to the WT level and restoration of twitching motility and adherence (Fig. 5A to C). The observation that the knh and pilT mutants have similar adherence levels suggests that both retractable and nonretractable pili mediate a basal level of adherence and that the pilT mutant is unable to utilize Knh as an adhesin. We next investigated if full-level adherence could be restored in the pilT mutant by surface capsule elimination via disruption of ctrA. As expected, the ctrA/pilT double mutant adhered to epithelial cells at the WT level (Fig. 5C). These data support the hypothesis that retraction of type IV pili is necessary to overcome the inhibitory influence of the capsule on Knh-mediated adherence.
FIG 5 .
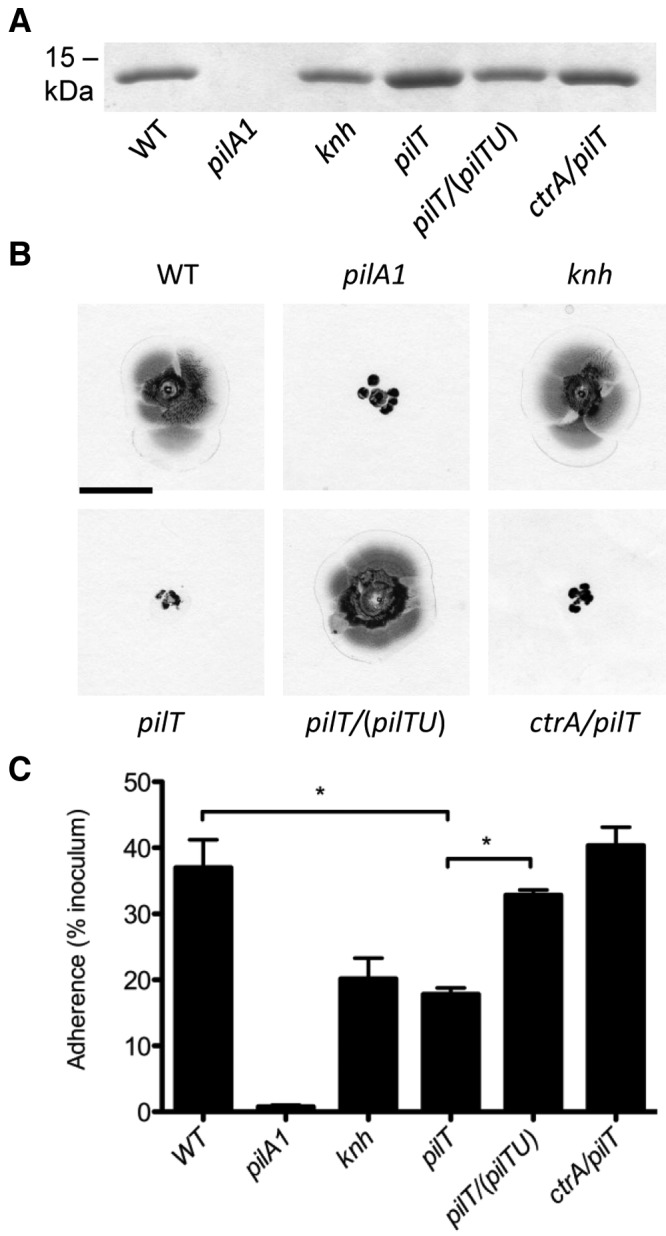
PilT/PilU retraction ATPase machinery influences K. kingae piliation, twitching motility, and adherence. K. kingae strains KK03, KK03pilA1, KK03knh, KK03pilT, KK03pilT/(pilTU), and KK03ctrA/pilT were examined for extracellular pilus expression by semiquantitative pilus preparation (A), twitching motility by a modified agar stab assay (scale bar, 1 cm) (B), and adherence to Chang human epithelial cells (C). Adherence assays were conducted in triplicate. Error bars indicate standard errors of the means. Statistical analyses were performed by using the unpaired t test to compare the adherence levels of two strains as follows: P < 0.05 (*) for KK03 versus KK03pilT and for KK03pilT versus KK03pilT/(pilTU).
DISCUSSION
The pathogenesis of K. kingae disease is thought to begin with colonization of the upper respiratory tract. Previous work established that type IV pili promote K. kingae adherence to human epithelial cells (13). In this study, we identified a novel K. kingae trimeric autotransporter protein called Knh that also mediates adherence to human epithelial cells. Further analysis revealed that K. kingae expresses a polysaccharide capsule that interferes with Knh adhesive activity when type IV pili are absent. We observed that type IV pili are important to overcome the polysaccharide capsule and facilitate Knh-mediated adherence in the presence of the PilT/PilU ATPase retraction machinery, presumably as a result of pilus retraction.
Trimeric autotransporters are integral outer membrane proteins and appear to be universally associated with adhesive activity (28). In our study, K. kingae derivatives expressing the trimeric autotransporter Knh but lacking type IV pili and a capsule were adherent, while those lacking Knh, type IV pili, and a capsule were nonadherent, providing strong evidence that Knh is an adhesin. Analysis of the Knh amino acid sequence revealed the presence of conserved domains possibly involved in adhesive activity. The N-terminal region of Knh is predicted to encode three clusters of β-roll degenerate repeats termed YadA-like head domains based on characterization of the Y. enterocolitica YadA trimeric autotransporter adhesin. Studies of YadA have demonstrated that the head domain is responsible for YadA-mediated adhesive activity (20, 29), raising the possibility that this domain is involved in Knh adhesive function as well. In addition, Knh has an ISneck2 domain that is similar to the ISneck1 domain identified in the adhesive regions of the H. influenzae Hia and Hsf adhesins (18). It is interesting to speculate regarding the location of the adhesive domain(s) of Knh relative to the outer membrane and the presence of a capsule. We have clearly demonstrated that a nonpiliated K. kingae mutant that expresses Knh and a capsule is nonadherent. One possible explanation for this phenotype is that the adhesive domain of Knh is buried within the capsule and is thus not accessible to the host cell receptor. The ISneck2 domain is proximal to the membrane anchor, suggesting that it is likely to be located close to the bacterial surface and may be masked by surface structures, including the polysaccharide capsule. The YadA-like head domains are located distally and are also presumably masked by the polysaccharide capsule, perhaps unless the capsule is displaced.
Encapsulation of bacterial pathogens has been shown to confer phenotypes important for survival on the host, including antiphagocytic activity and serum resistance. In this study, we report that K. kingae expresses a surface-associated polysaccharide capsule. According to current understanding, the pathogenesis of K. kingae disease begins with colonization of the upper respiratory tract, followed by hematogenous dissemination to joints, bones, or the endocardium (8). This pathogenic sequence is supported by the finding that genotypically identical K. kingae strains can be isolated from the pharynx and bloodstream of patients with systemic disease (12). We speculate that encapsulation is important for K. kingae to cause invasive disease by promoting intravascular survival, similar to observations with N. meningitidis, H. influenzae, and other encapsulated pathogens (30–34). Studies are under way to investigate the role of the K. kingae capsule in serum resistance and antiphagocytic activity. In addition to expressing a surface-associated polysaccharide capsule, K. kingae produces a secreted exopolysaccharide galactan that has broad antibiofilm activity (35). At this time, the relationship between the capsular polysaccharide and the galactan is unclear.
This study established that surface expression of the K. kingae capsule requires a predicted ABC-type transporter system with homology to CtrABCD in N. meningitidis. We also identified homologs of the N. meningitidis CtrE/LipA and CtrF/LipB proteins (36, 37). Both of these proteins also have homologs in the E. coli group 2 and 3 capsule biosynthesis systems (KpsC and KpsS) and in encapsulated H. influenzae (HcsA and HcsB) and appear to play a role in the export of capsule polymers to the bacterial surface (38, 39). Interestingly, the genetic organization of the polysaccharide synthesis machinery in K. kingae differs from that of E. coli groups 2 and 3, H. influenzae, and N. meningitidis. In N. meningitidis, a capsule synthesis operon (which differs in gene composition, depending on the specific capsule serotype) is located immediately upstream of the ctrABCD export operon and is divergently transcribed (40, 41). In E. coli group 2, the synthesis operon is located immediately between the export and assembly operons, and in H. influenzae, the synthesis operon is adjacent to the export operon (42–46). We have been unable to locate a putative capsule synthesis operon adjacent to ctrABCD or at any other locus in our draft K. kingae genome sequence, suggesting that K. kingae utilizes a unique genetic mechanism for the synthesis of capsular polysaccharide in a system that utilizes the ABC-type transporter system for export.
We first suspected that the capsule may be obstructing the ability of Knh to mediate adherence when we found that a mutant KK03 strain expressing a capsule and Knh but lacking pili was nonadherent. In considering this possibility, elaboration of a polysaccharide capsule has been shown to interfere with adhesive interactions with the host in several other pathogens. For example, the capsule interferes with adherence to human epithelial cells by H. influenzae type b (47), with adherence to gastrointestinal epithelium by enterotoxigenic E. coli (48), and with N. meningitidis Opa- and Opc-mediated adherence to epithelial and endothelial cells (49, 50). It is intriguing that the adherence of the ctrA mutant is slightly decreased compared that of parental strain KK03. While it is clear that the surface capsule blocks Knh-mediated adherence in the absence of type IV pili, we speculate that interaction of pili, Knh, and the capsule is necessary for full-level adherence of the WT organism. It is interesting to compare the K. kingae adherence data presented here with the adherence mechanism of N. meningitidis, a related encapsulated member of the Neisseriaceae family. N. meningitidis uses a two-step adherence process that starts with an initial type IV pilus-mediated interaction associated with microcolony formation on the host cell (51). Type IV pili are the key surface factor for adherence of encapsulated N. meningitidis to the epithelium and endothelium (52, 53). Following the initial adhesive interaction, type IV pili and the capsule are downregulated during the transition to the second stage of host cell interaction, which is termed intimate adherence (51, 54, 55). The shift from initial adherence to intimate adherence occurs over a course of hours, leading to host cell cytoskeletal rearrangements and cortical plaque formation under the intimately adherent bacteria (51, 56). Interestingly, we were able to detect high-level adherence after an only 25-min incubation of WT K. kingae with human epithelial cells, indicating that type IV pili are able to rapidly overcome the inhibitory influence of the capsule on Knh-mediated adherence, apparently via PilT/PilU-mediated retraction. We have observed similar adherence after a 5-min incubation (data not shown), which argues against the downregulation of capsule expression. These results suggest that K. kingae has developed a unique mechanism to efficiently adhere to host cells without having to downregulate capsule expression and become more susceptible to immune clearance.
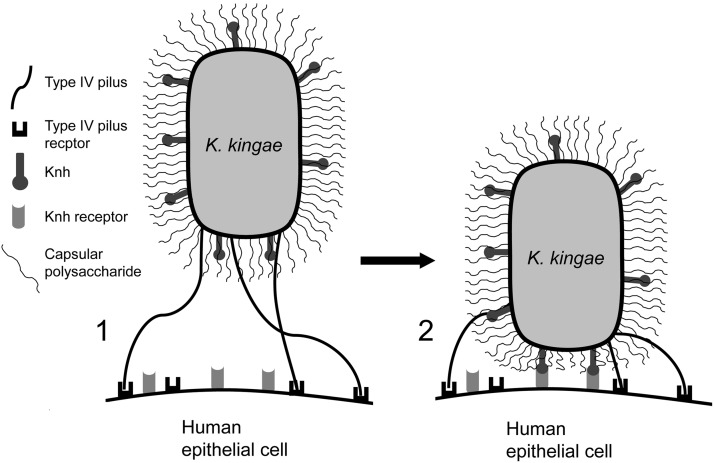
We propose a model for K. kingae adherence to human epithelial cells focusing on the three surface factors examined in this study: type IV pili, Knh, and the capsule (Fig. 6). This model involves a two-step process that begins with a low-affinity adhesive event mediated by type IV pili, as these fibers are able to extend beyond the polysaccharide capsule and engage their host cell receptor. PilT/PilU-mediated pilus retraction then brings the bacterium into close contact with the host cell, causing physical displacement of the surface polysaccharide capsule. Once the capsule has been displaced, Knh is able to engage its host cell receptor, leading to the high-level adherence we observe in adherence assays. It is important to note that this model relies on strong pilus retraction forces to displace the capsule. The force generated by a single Myxococcus xanthus type IV pilus has been measured at ~150 pN, making this the strongest linear molecular motor described to date (57). Similar measurements have been made for N. gonorrhoeae type IV pili, suggesting that the strong forces generated by these bacterial organelles may be conserved in the Neisseriaceae family (58).
FIG 6 .
K. kingae adherence to human epithelial cells involves sequential adhesive events. Initial K. kingae interactions with the host respiratory epithelium are initiated by type IV pili (1). Once pili have engaged their host cell receptor, PilT-mediated pilus retraction pulls the bacterium into close contact with the host cell membrane. This action results in physical displacement of the polysaccharide capsule, exposing Knh to the host cell membrane. Unmasked Knh can then mediate high-affinity K. kingae interactions with human epithelial cells via the Knh host cell receptor without the need for genetic capsule downregulation (2).
Our results establish a potential mechanism by which K. kingae adheres to the host respiratory epithelium and promotes colonization of the upper respiratory tract. However, the precise interrelationship and regulatory mechanisms of the three surface factors presented here have yet to be determined. Further investigation of the K. kingae adherence mechanism may lead to novel strategies for the prevention of disease due to K. kingae and to other encapsulated pathogenic bacteria.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table S2 in the supplemental material. Strain 269-492 forms two distinct colony morphologies on solid agar that are referred to as KK01 and KK03. KK01 is associated with stable low-level piliation, and KK03 is associated with stable high-level piliation. KK03 was used as the WT strain throughout this study to control for the piliation level. K. kingae strains were stored at −80°C in brain heart infusion broth with 30% glycerol. E. coli strains were stored at −80°C in Luria-Bertani (LB) broth with 15% glycerol. K. kingae strains were routinely cultured at 37°C with 5% CO2 on chocolate agar supplemented with 50 µg/ml kanamycin, 1 µg/ml erythromycin, or 2 µg/ml tetracycline, as appropriate. E. coli strains were routinely cultured at 37°C in LB broth or on LB agar supplemented with 100 µg/ml ampicillin, 50 µg/ml kanamycin, 500 µg/ml erythromycin, or 12.5 µg/ml tetracycline, as appropriate.
Genetic disruptions.
Gene disruptions were introduced into K. kingae by natural transformation (59). Following recovery of transformants by plating on selective medium, correct localization of the gene disruptions was confirmed by Southern blotting or PCR. The pilA1 gene was disrupted as described previously (13). All of the plasmids used in this study are listed in Table S2 in the supplemental material, and the sequences of all of the primers used are listed in Table S3. To disrupt the knh gene, fragments corresponding to the 5′ and 3′ regions of the gene were individually amplified from K. kingae strain 269-492 by using primers knh5′ F and knh5′ R and primers knh3′ F and knh3′ R, respectively. These fragments were ligated into BamHI/SalI-digested pTrc99A (introducing an internal deletion and a ClaI site within knh), generating pTrc99A/knh::ClaI. The tetM tetracycline resistance cassette was amplified with flanking ClaI sites from pHSXtetM4 using primers tetM F and tetM R and ligated into pTrc99A/knh::ClaI, generating pTrc99A/knh::tetM. To disrupt the ctrA gene, a fragment containing most of the ctrA ORF and the upstream flanking sequence was amplified from K. kingae strain 269-492 with primers ctrA F and ctrA R and ligated into BamHI/HindIII-digested pUC19, generating pUC19/ctrA. Subsequently, the QuikChange XL site-directed mutagenesis kit (Agilent Technologies) was used to insert an MluI site into the ctrA ORF by using primers ctrA MluI F and ctrA MluI R, generating pUC19/ctrA::MluI. The erythromycin resistance cassette ermC was amplified with flanking MluI sites from pIDN4 by using primers ermC F and ermC R and ligated into pUC19/ctrA::MluI, generating pUC19/ctrA::ermC. To disrupt the pilT gene, a fragment containing most of the pilT ORF and the upstream flanking sequence was amplified from K. kingae strain 269-492 with primers pilT F and pilT R and ligated into BamHI/HindIII-digested pUC19, generating pUC19/pilT. Subsequently, site-direct mutagenesis was used to insert an MluI site into the pilT ORF by using primers pilT MluI F and pilT MluI R, generating pUC19/pilT::MluI. The kanamycin resistance cassette aphA3 was excised from pFalcon2 by digestion with MluI and ligated into MluI-digested pUC19/pilT::MluI, generating pUC19/pilT::aphA3. To rule out PilA1 amino acid sequence variation as a source of phenotypic changes, the pilA1 gene in all strains that did not contain the pilA1 disruption was sequenced and confirmed to be 100% identical to the KK03 pilA1 sequence. To exclude possible phase-variable events in the mutants generated in this study, all strains were examined for pilus production by using extracellular pilus preparations, Knh production using Western blotting, and capsule production using heat extraction and alcian blue staining to confirm the presence or absence of the three surface factors under study.
The genetic complementation strategy used is detailed in Fig. S1 and Text S1 in the supplemental material.
Generation of anti-Knh polyclonal antiserum.
In order to generate an antiserum reactive with Knh, a 1.3-kb fragment corresponding to the 5′ coding region of knh (encoding amino acids 55 to 403, lacking the predicted signal sequence) was amplified with primers KnhAb F and KnhAb R and ligated into EcoRI/SalI-digested pGEX6p (GE Healthcare Life Sciences), creating pGEX::Knh55-403, which encodes a glutathione S-transferase (GST) fusion protein. The GST-Knh55-403 fusion protein was affinity purified on glutathione-agarose and treated with PreScission protease (GE Healthcare Life Sciences) to release Knh55-403 from the GST fusion. The resulting 42-kDa protein was resolved by 10% SDS-PAGE, stained with Coomassie blue, excised, and then injected into a guinea pig, generating antiserum GP97.
Protein analysis.
A 264-bp fragment encoding Knh amino acids 1695 to 1783 encompassing the predicted beta-barrel membrane anchor was PCR amplified with primers KnhBB F and KnhBB R, digested with EcoRI and BamHI, and ligated into EcoRI/BamHI-digested pASK-IBA12 (IBA BioTAGnologies). The resulting construct, designated pASK::Knh1695-1783, was transformed into E. coli BL21 omp8 and induced with 2 µg/ml anhydrous tetracycline for 3 h. Outer membranes were isolated on the basis of Sarkosyl insolubility, and an aliquot was treated with 95% formic acid overnight as previously described (21). Samples with or without formic acid treatment were separated by 15% SDS-PAGE and transferred to nitrocellulose. Western blot analysis of Strep-tagged recombinant KnhBB was performed by using the anti-Strep monoclonal antibody (IBA BioTAGnologies) according to the manufacturer’s recommendations.
Western blot analysis of Knh in K. kingae was performed as described previously, with Knh-specific antiserum (60). Briefly, outer membrane fractions were isolated on the basis of sarkosyl insolubility and treated overnight with 95% formic acid. Samples were resolved by 7.5% SDS-PAGE, transferred to nitrocellulose, and probed with antiserum GP97 against Knh and a secondary goat anti-guinea pig horseradish peroxidase-conjugated antibody. Western blot assays were developed by using a chemiluminescent peroxidase substrate.
Pilus preparations.
Derivatives of K. kingae strain KK03 were incubated on chocolate agar for 17 to 18 h, and growth was suspended in 1.5 ml 50 mM Tris–150 mM NaCl, pH 8.0, to an optical density at 600 nm (OD600) of 1.0, vortexed at full speed for 1 min, and centrifuged at 21,000 × g for 2 min to pellet the bacteria. A 1.25-ml volume of the bacterium-free supernatant was subjected to 20% ammonium sulfate precipitation on ice for 2 h. Precipitated pili were collected via centrifugation at 21,000 × g for 5 min and resuspended in 1× SDS-PAGE loading buffer. Aliquots were separated by 15% SDS-PAGE and stained with Coomassie blue. The observed protein band was confirmed to be the major pilin subunit, PilA1, by Western blot analysis with antiserum GP65 as described previously (14).
Capsule extraction.
Derivatives of K. kingae strain KK03 and H. influenzae type b strain C54 were incubated for 17 to 18 h on chocolate agar, suspended in 1 ml phosphate-buffered saline (PBS) to an OD600 of 0.8, centrifuged for 2 min at 10,000 × g, and resuspended in 0.5 ml PBS. Suspensions were incubated at 55°C for 30 min to extract capsular material. Bacteria were pelleted, and supernatants were concentrated 10-fold in an Amicon Ultra centrifuge filter with a 10,000 molecular weight cutoff. Capsule extracts were separated by 7.5% SDS-PAGE and stained with the cationic dye alcian blue (0.125% alcian blue in 40% ethanol/5% acetic acid; Sigma) for 2 h and then destained overnight in 40% ethanol/5% acetic acid.
Cationic ferritin staining.
Derivatives of K. kingae strain KK03 were incubated on chocolate agar for 17 to 18 h, and growth was suspended in 3 ml 0.1 M sodium cacodylate, pH 7.0 (cacodylate buffer), to an OD600 of 0.8. Glutaraldehyde was added to a final concentration of 5%, and samples were incubated for 20 min at ambient temperature. Following centrifugation at 750 × g for 15 min, pellets were gently resuspended in 1 ml cacodylate buffer, polycationic ferritin (Sigma) was added to a final concentration of 1 mg/ml, and samples were incubated for 30 min at ambient temperature and then centrifuged at 750 × g for 15 min. The pellets were washed once with 1 ml cacodylate buffer, resuspended in cacodylate buffer with 5% glutaraldehyde, incubated for 2 h at ambient temperature and then overnight at 4°C, and processed for TEM analysis.
TEM.
To assess surface capsule expression, cationic-ferritin-stained bacteria were washed three times with cacodylate buffer, embedded in 2% low-melting-point agarose, and fixed in cacodylate buffer with 4% glutaraldehyde and 7.5% sucrose. The pellets were postfixed in 1% osmium tetroxide in cacodylate buffer, stained en bloc with 0.5% uranyl acetate, dehydrated with a graded series of ethanol, and embedded in epoxy resin. Blocks were cut into thin sections (60 to 90 nm), placed on copper-rhodium 200-mesh grids, and examined by using a Philips CM-12 electron microscope.
Twitching motility assays.
Twitching motility was assessed by a modified agar plate stab assay originally developed for P. aeruginosa (27). Briefly, derivatives of K. kingae strain KK03 were incubated on chocolate agar for 17 to 18 h and growth was resuspended in 1× PBS to an OD600 of 1.0. One microliter of the bacterial suspension was stab inoculated into the bottom of tissue culture-treated 100-mm plates containing chocolate agar and incubated at 37°C with 5% CO2 for 2 days. Twitching motility-competent strains spread from the stab inoculation site at the plate-agar interface. The agar was carefully peeled away, and the plate was air dried and stained with crystal violet to visualize the twitching motility zones.
Eukaryotic cell lines and adherence.
Chang cells (Wong-Kilbourne derivative [D] of Chang conjunctiva, HeLa origin; ATCC CCL-20.2) were cultivated at 37°C with 5% CO2 in medium as described previously (13). Quantitative adherence assays were performed as described previously (13). Briefly, confluent and fixed cell monolayers in 24-well plates were inoculated with approximately 6.5 × 106 CFU of bacteria and the plates were centrifuged at 165 × g for 5 min and then incubated for 25 min at 37°C in 5% CO2. Monolayers were washed four times with PBS to remove nonadherent bacteria and then treated with 1× trypsin-EDTA (Sigma) for 20 min at 37°C to release adherent bacteria. Percent adherence was calculated by dividing the number of adherent CFU by the number of inoculated CFU. All experiments were performed in triplicate. Statistical analysis was performed using the unpaired t test to compare the adherence levels of two strains as noted in the figure legends.
Nucleotide sequence accession numbers.
The nucleotide sequences of the ctrABCD, ctrE, ctrF, and knh loci have been submitted to GenBank and assigned accession numbers JX944815 (ctrABCD), JX944816 (ctrE), JX944817 (ctrF), and JX944818 (knh).
SUPPLEMENTAL MATERIAL
Supplemental methods. Download Text S1, PDF file, 0.1 MB.
Predicted amino acid homology between K. kingae strain 296-492 and N. meningitidis MC58 Ctr proteins.
Strains and plasmids used in this study.
Primers used in this study.
Schematic representation of the K. kingae chromosomal complementation construct. The pUC19 vector backbone contains the 5′ and 3′ homologous targeting regions to direct chromosomal insertion, the ermC erythromycin resistance marker (Ermr), and a portion of the pUC19 multiple cloning site for insertion of the desired gene(s) for complementation. After the desired gene(s) is ligated into the complementation construct, the pUC19 vector backbone is linearized with a single-cutter restriction enzyme and transformed into K. kingae. Download Figure S1, TIF file, 0.3 MB.
Mutation of ctrA does not have a polar effect on bccA. (A) Diagram of the WT ctrABCD locus in K. kingae with the downstream gene, bccA. The ctrA gene encodes a predicted outer membrane polysaccharide export pore, ctrB encodes a predicted inner membrane- and periplasm-spanning polysaccharide copolymerase, and ctrC and ctrD encode a predicted ATP-dependent (ABC-type) transport cassette. (B) qRT-PCR analysis of bccA, encoding a predicted BCCT family betaine/carnitine/choline transporter, of strain KK03ctrA compared to that of KK03. Download Figure S2, TIF file, 0.1 MB.
ACKNOWLEDGMENTS
We thank Sara Miller of the Duke Electron Microscopy Service for assistance with TEM and Hank Seifert for plasmids pHSXtetM4 and pIDN4.
Footnotes
Citation:Porsch EA, Kehl-Fie TE, St. Geme JW, III. 2012. Modulation of Kingella kingae adherence to human epithelial cells by type IV pili, capsule, and a novel trimeric autotransporter. mBio 3(5):e00372-12. doi:10.1128/mBio.00372-12.
REFERENCES
- 1. Chometon S, et al. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26:377–381 [DOI] [PubMed] [Google Scholar]
- 2. Gené A, García-García JJ, Sala P, Sierra M, Huguet R. 2004. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr. Infect. Dis. J. 23:886–888 [DOI] [PubMed] [Google Scholar]
- 3. Lehours P, et al. 2011. The rtxA toxin gene of Kingella kingae: a pertinent target for molecular diagnosis of osteoarticular infections. J. Clin. Microbiol. 49:1245–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moumile K, Merckx J, Glorion C, Berche P, Ferroni A. 2003. Osteoarticular infections caused by Kingella kingae in children: contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 22:837–839 [DOI] [PubMed] [Google Scholar]
- 5. Verdier I, et al. 2005. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr. Infect. Dis. J. 24:692–696 [DOI] [PubMed] [Google Scholar]
- 6. Yagupsky P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4:358–367 [DOI] [PubMed] [Google Scholar]
- 7. Yagupsky P, et al. 1992. High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J. Clin. Microbiol. 30:1278–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yagupsky P, Porsch E, St Geme JW., III 2011. Kingella kingae: an emerging pathogen in young children. Pediatrics 127:557–565 [DOI] [PubMed] [Google Scholar]
- 9. Yagupsky P, Dagan R, Prajgrod F, Merires M. 1995. Respiratory carriage of Kingella kingae among healthy children. Pediatr. Infect. Dis. J. 14:673–678 [DOI] [PubMed] [Google Scholar]
- 10. Yagupsky P, et al. 2009. Dissemination of Kingella kingae in the community and long-term persistence of invasive clones. Pediatr. Infect. Dis. J. 28:707–710 [DOI] [PubMed] [Google Scholar]
- 11. Yagupsky P, Peled N, Katz O. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 40:4180–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yagupsky P, Porat N, Pinco E. 2009. Pharyngeal colonization by Kingella kingae in children with invasive disease. Pediatr. Infect. Dis. J. 28:155–157 [DOI] [PubMed] [Google Scholar]
- 13. Kehl-Fie TE, Miller SE, St Geme JW., III 2008. Kingella kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells. J. Bacteriol. 190:7157–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW., III 2009. Expression of Kingella kingae type IV pili is regulated by sigma54, PilS, and PilR. J. Bacteriol. 191:4976–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kehl-Fie TE, et al. 2010. Examination of type IV pilus expression and pilus-associated phenotypes in Kingella kingae clinical isolates. Infect. Immun. 78:1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peak IR, Srikhanta Y, Dieckelmann M, Moxon ER, Jennings MP. 2000. Identification and characterisation of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 28:329–334 [DOI] [PubMed] [Google Scholar]
- 17. Scarselli M, et al. 2006. Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61:631–644 [DOI] [PubMed] [Google Scholar]
- 18. Meng G, St Geme JW, III, Waksman G. 2008. Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. J. Mol. Biol. 384:824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nummelin H, et al. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23:701–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tahir YE, Kuusela P, Skurnik M. 2000. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG–S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 37:192–206 [DOI] [PubMed] [Google Scholar]
- 21. St Geme JW, III, Kumar VV, Cutter D, Barenkamp SJ. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karlyshev AV, McCrossan MV, Wren BW. 2001. Demonstration of polysaccharide capsule in Campylobacter jejuni using electron microscopy. Infect. Immun. 69:5921–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pujol C, Eugène E, Marceau M, Nassif X. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A. 96:4017–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitchurch CB, Hobbs M, Livingston SP, Krishnapillai V, Mattick JS. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33–44 [DOI] [PubMed] [Google Scholar]
- 26. Wolfgang M, et al. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321–330 [DOI] [PubMed] [Google Scholar]
- 27. Alm RA, Mattick JS. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485–496 [DOI] [PubMed] [Google Scholar]
- 28. Łyskowski A, Leo JC, Goldman A. 2011. Structure and biology of trimeric autotransporter adhesins. Adv. Exp. Med. Biol. 715:143–158 [DOI] [PubMed] [Google Scholar]
- 29. Tamm A, et al. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995–1011 [DOI] [PubMed] [Google Scholar]
- 30. Kahler CM, et al. 1998. The (alpha2->8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect. Immun. 66:5939–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mackinnon FG, et al. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb. Pathog. 15:359–366 [DOI] [PubMed] [Google Scholar]
- 32. Vogel U, Hammerschmidt S, Frosch M. 1996. Sialic acids of both the capsule and the sialylated lipooligosaccharide of Neisseria meningitis serogroup B are prerequisites for virulence of meningococci in the infant rat. Med. Microbiol. Immunol. 185:81–87 [DOI] [PubMed] [Google Scholar]
- 33. Moxon ER, Vaughn KA. 1981. The type b capsular polysaccharide as a virulence determinant of Haemophilus influenzae: studies using clinical isolates and laboratory transformants. J. Infect. Dis. 143:517–524 [DOI] [PubMed] [Google Scholar]
- 34. Zwahlen A, Kroll JS, Rubin LG, Moxon ER. 1989. The molecular basis of pathogenicity in Haemophilus influenzae: comparative virulence of genetically-related capsular transformants and correlation with changes at the capsulation locus cap. Microb. Pathog. 7:225–235 [DOI] [PubMed] [Google Scholar]
- 35. Bendaoud M, et al. 2011. Broad-spectrum biofilm inhibition by Kingella kingae exopolysaccharide. J. Bacteriol. 193:3879–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frosch M, Müller A. 1993. Phospholipid substitution of capsular polysaccharides and mechanisms of capsule formation in Neisseria meningitidis. Mol. Microbiol. 8:483–493 [DOI] [PubMed] [Google Scholar]
- 37. Tzeng YL, et al. 2005. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect. Immun. 73:1491–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bliss JM, Silver RP. 1996. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli. Mol. Microbiol. 21:221–231 [DOI] [PubMed] [Google Scholar]
- 39. Sukupolvi-Petty S, Grass S, St Geme JW., III 2006. The Haemophilus influenzae type b hcsA and hcsB gene products facilitate transport of capsular polysaccharide across the outer membrane and are essential for virulence. J. Bacteriol. 188:3870–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swartley JS, Ahn JH, Liu LJ, Kahler CM, Stephens DS. 1996. Expression of sialic acid and polysialic acid in serogroup B Neisseria meningitidis: divergent transcription of biosynthesis and transport operons through a common promoter region. J. Bacteriol. 178:4052–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. 1991. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol. Microbiol. 5:1251–1263 [DOI] [PubMed] [Google Scholar]
- 42. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 43. Whitfield C, Roberts IS. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307–1319 [DOI] [PubMed] [Google Scholar]
- 44. Kroll JS, Loynds B, Brophy LN, Moxon ER. 1990. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol. Microbiol. 4:1853–1862 [DOI] [PubMed] [Google Scholar]
- 45. Kroll JS, Zamze S, Loynds B, Moxon ER. 1989. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J. Bacteriol. 171:3343–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Eldere J, et al. 1995. Region II of the Haemophilus influenzae type be capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107–118 [DOI] [PubMed] [Google Scholar]
- 47. St Geme JW, III, Falkow S. 1991. Loss of capsule expression by Haemophilus influenzae type b results in enhanced adherence to and invasion of human cells. Infect. Immun. 59:1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Runnels PL, Moon HW. 1984. Capsule reduces adherence of enterotoxigenic Escherichia coli to isolated intestinal epithelial cells of pigs. Infect. Immun. 45:737–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Virji M, Makepeace K, Ferguson DJ, Achtman M, Moxon ER. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10:499–510 [DOI] [PubMed] [Google Scholar]
- 50. Virji M, et al. 1992. Expression of the Opc protein correlates with invasion of epithelial and endothelial cells by Neisseria meningitidis. Mol. Microbiol. 6:2785–2795 [DOI] [PubMed] [Google Scholar]
- 51. Pujol C, Eugène E, de Saint Martin L, Nassif X. 1997. Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun. 65:4836–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Virji M, Alexandrescu C, Ferguson DJ, Saunders JR, Moxon ER. 1992. Variations in the expression of pili: the effect on adherence of Neisseria meningitidis to human epithelial and endothelial cells. Mol. Microbiol. 6:1271–1279 [DOI] [PubMed] [Google Scholar]
- 53. Virji M, et al. 1991. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol. Microbiol. 5:1831–1841 [DOI] [PubMed] [Google Scholar]
- 54. Deghmane AE, Giorgini D, Larribe M, Alonso JM, Taha MK. 2002. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 43:1555–1564 [DOI] [PubMed] [Google Scholar]
- 55. Deghmane AE, et al. 2000. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 19:1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Merz AJ, Enns CA, So M. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32:1316–1332 [DOI] [PubMed] [Google Scholar]
- 57. Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. 2009. High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191:4633–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maier B, et al. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U. S. A. 99:16012–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kehl-Fie TE, St Geme JW., III 2007. Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J. Bacteriol. 189:430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sheets AJ, Grass SA, Miller SE, St Geme JW., III 2008. Identification of a novel trimeric autotransporter adhesin in the cryptic genospecies of Haemophilus. J. Bacteriol. 190:4313–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Text S1, PDF file, 0.1 MB.
Predicted amino acid homology between K. kingae strain 296-492 and N. meningitidis MC58 Ctr proteins.
Strains and plasmids used in this study.
Primers used in this study.
Schematic representation of the K. kingae chromosomal complementation construct. The pUC19 vector backbone contains the 5′ and 3′ homologous targeting regions to direct chromosomal insertion, the ermC erythromycin resistance marker (Ermr), and a portion of the pUC19 multiple cloning site for insertion of the desired gene(s) for complementation. After the desired gene(s) is ligated into the complementation construct, the pUC19 vector backbone is linearized with a single-cutter restriction enzyme and transformed into K. kingae. Download Figure S1, TIF file, 0.3 MB.
Mutation of ctrA does not have a polar effect on bccA. (A) Diagram of the WT ctrABCD locus in K. kingae with the downstream gene, bccA. The ctrA gene encodes a predicted outer membrane polysaccharide export pore, ctrB encodes a predicted inner membrane- and periplasm-spanning polysaccharide copolymerase, and ctrC and ctrD encode a predicted ATP-dependent (ABC-type) transport cassette. (B) qRT-PCR analysis of bccA, encoding a predicted BCCT family betaine/carnitine/choline transporter, of strain KK03ctrA compared to that of KK03. Download Figure S2, TIF file, 0.1 MB.