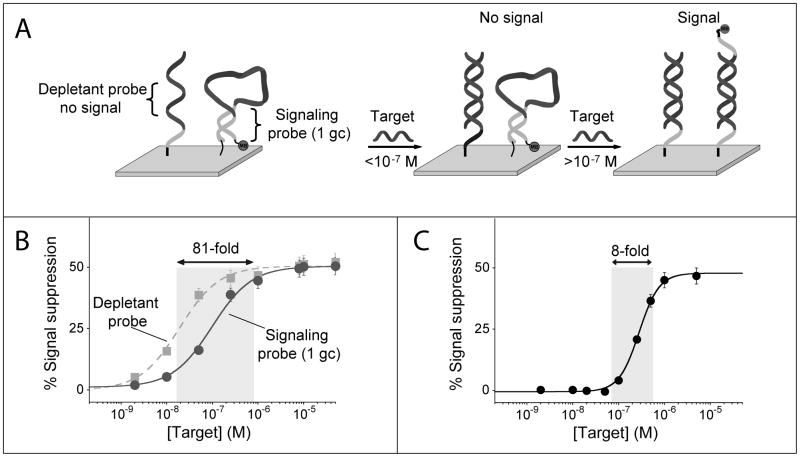
Figure 3.
Using the sequestration mechanism we can dramatically narrow the useful dynamic range of an E-DNA sensor, thus greatly improving its sensitivity (i.e., its ability to measure small changes in target concentration). (A) We do so by co-immobilizing on a single electrode surface a low affinity, signaling E-DNA probe with a higher affinity probe (depletant) which, lacking the redox reporter, does not signal upon binding its target. At low concentrations the target preferentially binds the depletant, which removes (sequesters) target from the sample without generating a signal. When the total target amount surpasses that of the depletant (the sink is saturated), a threshold response is achieved in which further addition of target dramatically raises the relative concentration of free target. This gives rise to a much steeper dose-response curve than this would occur in the absence of a depletant. (C) Using this approach we have narrowed the 81-fold useful dynamic range of an unmodified E-DNA sensor to a mere 8-fold, thus increasing its sensitivity by an order of magnitude.