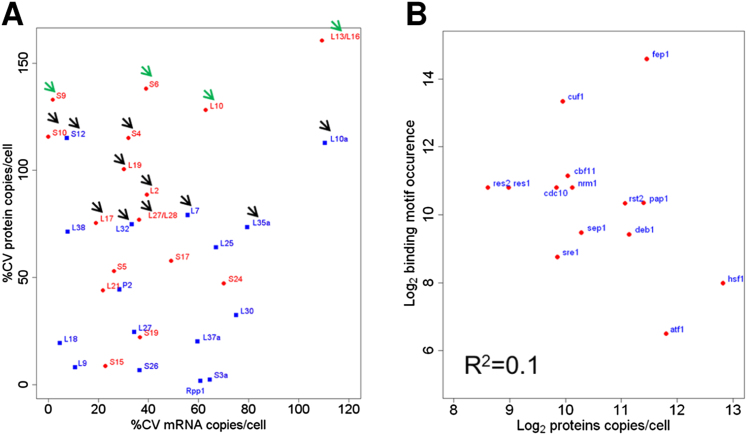
Figure S6.
Additional Analyses on Ribosomal Protein Paralogs and Transcription Factors, Related to Figure 5
(A) Expression variability of ribosomal proteins. Percent coefficient of variation in mRNA and protein expression of paralog genes from different ribosomal proteins families. Red dots: families containing repeated sequences (ambiguous mapping by RNA-seq). Blue squares: families with non-repeated sequences (un-ambiguous mapping by RNA-seq). Dots are labeled with ribosomal protein family. Black arrows: families with over 3-fold difference in protein expression between lowest and highest expressed member. Green arrows: as black arrows but for 10-fold difference.
The median mRNA expression of single-copy ribosomal proteins is significantly higher than the median mRNA expression of duplicated ribosomal proteins (1.7-fold, Pwilcox < 10−4). This patterns holds also for protein expression (1.7-fold, Pwilcox < 0.02). As most paralogs are found in two copies, this finding suggests that each paralog might contribute to about half of the ribosomes. Fission yeast ribosomes could therefore be heterogeneous complexes with respect to paralogs. However, possible technical or classification artifacts could contribute to this observation, as RNA-seq and the proteomics approach cannot unambiguously assign expression levels to paralogs with almost identical sequences. To look in more detail into paralog expression, we calculated the percent coefficient of variation (%CV) of mRNA and protein expression for each ribosomal protein family. High %CV indicates large differences in expression between paralogs. This analysis indicates that families with low %CV, where both paralogs are likely to contribute to ribosomes, can be found in cases where sequences were sufficiently diverged to permit reliable read assignment (blue squares). Moreover, some families showed vastly divergent protein expression between paralogs (arrows), suggesting either the existence of rare specialized ribosomes or extra-ribosomal functions of the lowly expressed paralogs.
(B) Comparison of TF expression levels with the occurrence of their DNA-binding motifs in the genome. Expression levels of TFs for which the DNA-binding motifs are available in PomBase were plotted against the number of their respective motifs found in the genome. Each dot represents a TF with its common name indicated in blue. It is problematic to localize true TF binding sites based on genome sequence alone, and ChIP-chip or ChIP-seq data are required to identify accurately the number of functionally relevant motifs.