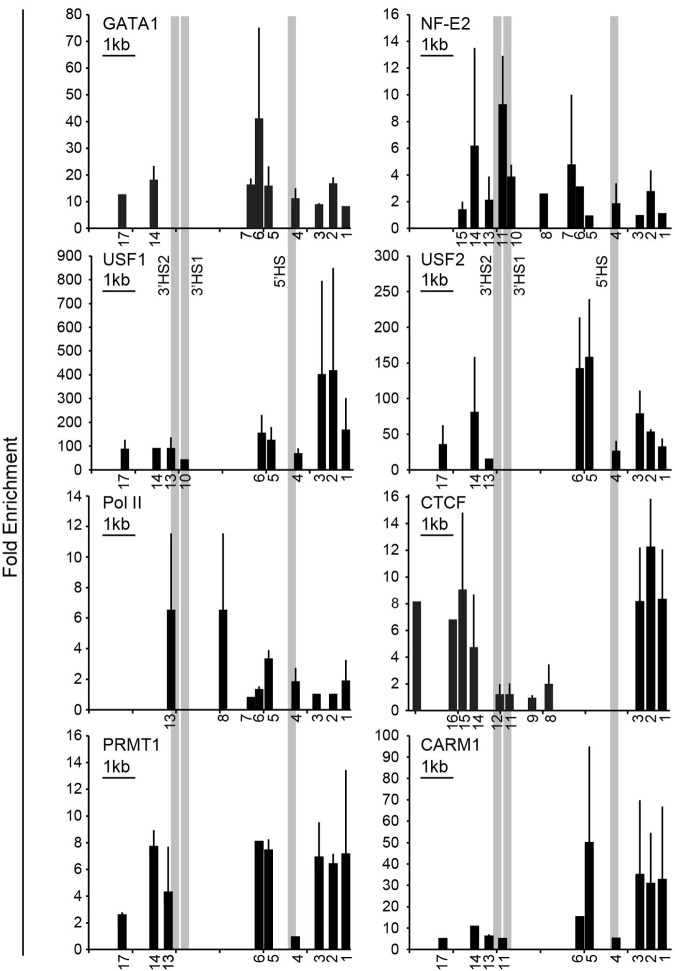
Figure 3.
Validation of DNA binding protein occupancy. ChIP analysis of K562 chromatin for occupancy by the erythroid-specific transcription factors GATA1 and NF-E2; RNA Pol II; the chromatin architectural protein CTCF; and the barrier-associated proteins USF1, USF2, PRMT1, and CARM1. The numbers below the x-axes indicate the locations of the primers used (see supplemental Table 1 for coordinates) and are distributed to scale. A 1-kb scale bar is shown in the top left corner of each graph. The absence of a number below the x-axis indicates that the region either was not tested or the result was not significant. The y-axis shows the fold enrichment over precipitation with immunoglobulin G. The bars represent the results of multiple ChIP experiments. Each ChIP experiment was done in triplicate, and the standard deviation is displayed. Each bar shown is significant at P < .05 or lower compared with control. The CTCF results are the combination of several ChIP experiments and the relative enrichment cannot be compared.