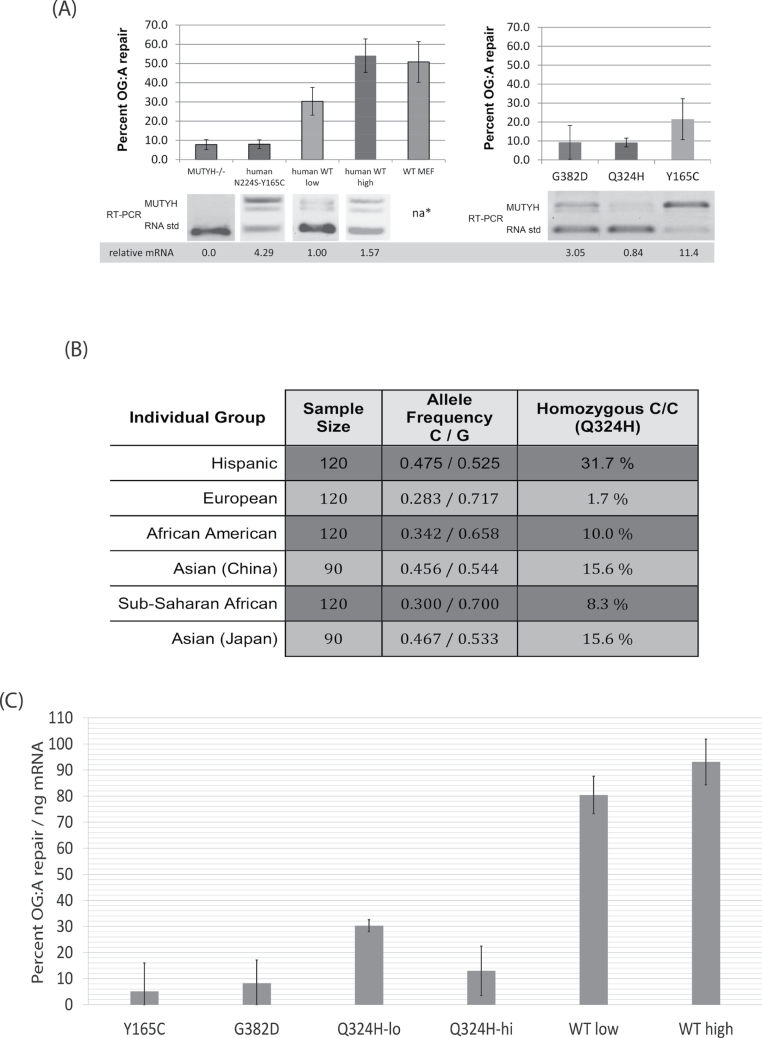
Fig. 3.
(A) Summary of flow cytometry results with the OG:A-containing fluorescent reporter and competitive RT–PCR of MUTYH expression in the corresponding MEF cell lines. Error bars represent the standard deviation from at least three separate experiments. na* endogenous mouse Mutyh not detected by RT–PCR due to differences in the gene sequence between species; western blot (Figure 1C) shows protein expression of WT MEFs is similar to the stable cell lines. GFP repair of the WT-lo cell line is significantly different from all other cell lines (P < 0.05). Also see Supplementary Table 1, available at Carcinogenesis Online) (B) Evidence that the SNP corresponding to the Q324H variant is common in diverse human groups. Matched subset of NCBI data for SNP rs3219489, format adapted from dbSNP (28) (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=3219489) collected by the Environmental Genome Project (54). The minor C allele encodes histidine (CAC), leading to the Q324H protein variant, whereas the major G allele encodes glutamine (CAG). Right: The frequency of homozygous carriers of Q324H in these samples. The minor allele frequency of all NCBI recorded samples is 31.9%. (C) Repair of OG:A (as measured by GFP fluorescence) normalized to MUTYH mRNA levels (as measured by competitive RT–PCR). Error bars are the square root of the sum of the variances from the RT–PCR and GFP fluorescence data, thus account for variation from both these data sources.