Abstract
Background
Transitions in habitats and feeding behaviors were fundamental to the diversification of life on Earth. There is ongoing debate regarding the typical directionality of transitions between aquatic and terrestrial habitats and the mechanisms responsible for the preponderance of terrestrial to aquatic transitions. Snail-killing flies (Diptera: Sciomyzidae) represent an excellent model system to study such transitions because their larvae display a range of feeding behaviors, being predators, parasitoids or saprophages of a variety of mollusks in freshwater, shoreline and dry terrestrial habitats. The remarkable genus Tetanocera (Tetanocerini) occupies five larval feeding groups and all of the habitat types mentioned above. This study has four principal objectives: (i) construct a robust estimate of phylogeny for Tetanocera and Tetanocerini, (ii) estimate the evolutionary transitions in larval feeding behaviors and habitats, (iii) test the monophyly of feeding groups and (iv) identify mechanisms underlying sciomyzid habitat and feeding behavior evolution.
Results
Bayesian inference and maximum likelihood analyses of molecular data provided strong support that the Sciomyzini, Tetanocerini and Tetanocera are monophyletic. However, the monophyly of many behavioral groupings was rejected via phylogenetic constraint analyses. We determined that (i) the ancestral sciomyzid lineage was terrestrial, (ii) there was a single terrestrial to aquatic habitat transition early in the evolution of the Tetanocerini and (iii) there were at least 10 independent aquatic to terrestrial habitat transitions and at least 15 feeding behavior transitions during tetanocerine phylogenesis. The ancestor of Tetanocera was aquatic with five lineages making independent transitions to terrestrial habitats and seven making independent transitions in feeding behaviors.
Conclusions
The preponderance of aquatic to terrestrial transitions in sciomyzids goes against the trend generally observed across eukaryotes. Damp shoreline habitats are likely transitional where larvae can change habitat but still have similar prey available. Transitioning from aquatic to terrestrial habitats is likely easier than the reverse for sciomyzids because morphological characters associated with air-breathing while under the water's surface are lost rather than gained, and sciomyzids originated and diversified during a general drying period in Earth's history. Our results imply that any animal lineage having aquatic and terrestrial members, respiring the same way in both habitats and having the same type of food available in both habitats could show a similar pattern of multiple independent habitat transitions coincident with changes in behavioral and morphological traits.
Background
Some of the most important evolutionary innovations in the history of life on Earth resulted from transitions between aquatic (freshwater) and terrestrial habitats. The colonization of land by unicellular aquatic plants [1,2] and their eventual transformation into vascular plants helped shape terrestrial environments and paved the way for the evolution of the majority of the eukaryotic species alive today. Other key lineage diversifications that occurred following transitions from aquatic to terrestrial habitats include those of tetrapod vertebrates [3], millipedes [4], scorpions [5], other arachnids [2,6,7], earth worms [8] and nematodes [9]. Whereas the ancestral insect originated in a terrestrial environment [10-12], insects are one of the most successful colonizers of freshwater habitats, as at least 12 of the 31 insect orders have representatives occupying these environments during at least one life history stage [13]. Transitions between aquatic and terrestrial habitats are generally rarer than other habitat changes (e.g., between epigeal and arboreal) because of the substantial physical differences between them [14]. In addition to differences in the physical requirements of living in water versus on land (e.g., differences in oxygen concentration), one presumed barrier is that the suite of available food items are typically distinct, as there are major differences between aquatic and terrestrial food webs [15,16]. Therefore, in order to transition between these habitats, a lineage typically must adapt to new physical conditions while concomitantly modifying its feeding behaviors.
The family Sciomyzidae, or “snail-killing flies” (Diptera: Acalyptratae: Sciomyzoidea), is an ideal taxon with which to study the evolution of feeding behaviors and associated habitat transitions. Their life histories are well-studied, as 240 of the 539 species have known larval feeding habits [17]. Sciomyzid larvae display a wide range of feeding behaviors, including predation, parasitism, or saprophagy of terrestrial, semi-aquatic and aquatic non-operculate snails, operculate aquatic snails, semi-terrestrial succineid snails, slugs, snail eggs, fingernail clams and freshwater oligochaete worms [17,18]. This represents, by far, the most extensive radiation of primarily malacophagous (= mollusk-feeding) species when compared to all other dipteran lineages [17,18]. A total of 109 species from six other dipteran families attack mollusks [19], whereas ~99% of the 240 sciomyzid species with known life cycles attack mollusks [17,18,20]. Sciomyzids have three larval stages and most species exhibiting parasitoid behavior have very specific host requirements in the 1st and 2nd larval stage but become more generalized predators in the 3rd stage. These species have been referred to as parasitoids or parasitoids/predators in sciomyzid literature, so, for simplicity, we refer to these species as parasitoids herein. There have been two different approaches to organizing sciomyzid species into behavioral/ecological groups: (i) based on commonalities in larval microhabitat, mode of feeding and prey type ([21]: 8 groups; [22]: 10 groups; [20]: 9 groups; [17,18]: 15 groups), and (ii) based on an ordination analysis of 36 egg and larval morphological characters, larval behaviors, and habitat that identified nine "Eco-Groups," each possessing a unique combination of states from these 36 characters [23].
The Sciomyzidae includes three subfamilies: the Huttonininae with two genera [24], the Salticellinae with one genus (Salticella) and the Sciomyzinae with the remaining 58 genera. The Sciomyzinae is comprised of two tribes, the Sciomyzini with 12 genera and the Tetanocerini with the remaining 46 genera [17]. All of the Sciomyzini and Salticellinae have terrestrial larvae, whereas 14 tetanocerine genera have at least one species with aquatic larvae [17]. The larvae of the Huttonininae remain unknown [17]. Recent phylogenetic analyses of morphological data suggest that the Sciomyzinae and its two tribes are monophyletic [23,25]. The family Phaeomyiidae, with five described Palaearctic species in two genera (Akebono and Pelidnoptera), was at one time considered to be a subfamily of the Sciomyzidae, but was subsequently elevated to family by Griffiths [26], who proposed its sister status to Sciomyzidae.
The evolution of feeding behaviors in Sciomyzidae has been discussed in numerous papers (e.g., [17,18,20-22]). Because larval feeding on decaying animal matter occurs in other dipteran lineages, including families in the Sciomyzoidea (e.g., Dryomyzidae; [17]), it has been suggested that the ancestral sciomyzid was probably similar to the extant Atrichomelina pubera (Sciomyzini), a generalist that feeds on dead, dying or living aquatic and semi-aquatic, non-operculate snails on damp terrestrial substrates [27,28]. Steyskal’s [29] classification of the Sciomyzidae lead to sciomyzine larvae being characterized as terrestrial (including those inhabiting moist surfaces) saprophages/predators/parasitoids, while tetanocerine larvae are typically characterized as aquatic predators. Knutson & Vala [18] mapped their feeding groups onto the morphological phylogeny presented in Marinoni & Mathis [25] to infer the ancestral feeding behavior for the family and to discuss the evolution of such behaviors based on the position of each genus in the phylogeny. They concluded that while Steyskal's [29] generalizations have exceptions, the distribution of feeding behaviors known today support these general characterizations. They further concluded that the terrestrial habits of many of the species in the Tetanocerini represent a derived condition within the tribe. Unfortunately, the utility of Knutson and Vala's [18] study was somewhat limited due to the incomplete resolution of intergeneric relationships and the absence of replicate intrageneric taxon sampling within the Marinoni & Mathis [25] phylogeny. A more recent study on intergeneric sciomyzid relationships [23], which included more morphological characters than did Marinoni & Mathis [25], was similarly limited, as the taxon sampling was nearly identical and the relationships among many of the genera not well-supported. Therefore, a well-resolved species-level phylogeny focusing on a lineage that exhibits a variety of feeding behaviors and occupies multiple habitats would enable a better understanding of the evolutionary processes involved in transitions among habitat, mode of feeding and host/prey selection.
Within the Tetanocerini, the genus Tetanocera is of particular interest because it is one of the most diverse sciomyzid genera with respect to feeding behaviors. Twenty-eight of its 39 species have known life cycles (see Table 1 for a partial list) and its species occupy five of the 15 feeding groups of Knutson & Vala [17,18]: (i) general predators of non-operculate aquatic snails in the water (14 species), (ii) general predators of non-operculate aquatic snails occurring on damp shorelines (3 species), (iii) general predators of terrestrial snails (2 species), (iv) parasitoids of slugs (4 species) or (v) parasitoids of succineid (semi-terrestrial) snails (5 species). The life cycles of 11 species remain unknown. Members of the largest feeding group within Tetanocera (i above) spend their larval stages just under the surface of the water, whereas the remaining groups have terrestrial larvae. Only one other sciomyzid genus occupies five feeding groups (Sepedon), whereas most only occupy one or two [17].
Table 1.
Species analyzed in this study, the feeding behavioral group[17]to which each taxon belongs, and GenBank numbers for the sequences used in this study
|
Family |
Genus |
Feeding Group |
Specimen |
GenBank Accession Numbers |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Tribe | Species | Feeding Group | Citation | Number | COI | COII | 16S | 28S | Ef-1α |
| Drosophilidae |
Drosophila melanogaster Meigen 1830 |
Yeast, mold |
[80] |
|
AJ400907 |
AJ400907 |
AJ400907 |
M21017 |
NM_170570 |
| Phaeomyiidae |
Pelidnoptera nigripennis (Fabricius 1794) |
Millipede parasitoid |
[81] |
F272 |
JN860439 |
|
|
JN837497 |
JN816249 |
| Sciomyzidae |
|
|
|
|
|
|
|
|
|
| Sciomyzini |
Atrichomelina |
|
|
|
|
|
|
|
|
| |
Atrichomelina pubera (Loew 1862) |
Facultative predator/saprophage of snails and clams on damp shorelines |
[28] |
F160 |
JN860438 |
JN837567 |
JN816281 |
JN837498 |
|
| |
|
|
|
F161 |
AY875151 |
AY875182 |
AY875089 |
AY875120 |
JN816247 |
| |
Sciomyza |
|
|
|
|
|
|
|
|
| |
Sciomyza simplex Fallén 1820 |
Predator of shoreline-stranded aquatics |
[82] |
F175 |
AY875152 |
AY875183 |
AY875090 |
AY875121 |
JN816248 |
| Tetanocerini |
Anticheta |
|
|
|
|
|
|
|
|
| |
Anticheta melanosoma Melander 1920 |
Predator of exposed snail eggs |
[83] |
F254 |
JN860440 |
JN837568 |
JN816327 |
JN837499 |
JN816250 |
| |
Dichetophora |
|
|
|
|
|
|
|
|
| |
Dichetophora finlandica Verbèke 1964 |
Unknown |
|
F248 |
JN860441 |
JN837569 |
JN816328 |
JN837500 |
JN816251 |
| |
Dictya |
|
|
|
|
|
|
|
|
| |
Dictya borealis Curran 1932 |
Predator of aquatic snails in the water |
[84] |
F257 |
JN860442 |
JN837570 |
JN816329 |
JN837501 |
JN816252 |
| |
Dictya expansa Steyskal 1938 |
Predator of aquatic snails in the water |
[84] |
F263 |
JN860443 |
JN837571 |
JN816330 |
JN837502 |
|
| |
Dictya floridensis Steyskal 1954 |
Predator of aquatic snails in the water |
[85] |
F258 |
JN860444 |
JN837572 |
JN816331 |
JN837503 |
|
| |
Dictya gaigei Steyskal 1938 |
Predator of aquatic snails in the water |
[84] |
F267 |
JN860445 |
JN837573 |
JN816336 |
JN837504 |
|
| |
Dictya pictipes (Loew 1859) |
Predator of aquatic snails in the water |
[84] |
F261 |
JN860446 |
JN837574 |
JN816332 |
JN837505 |
JN816253 |
| |
Dictya steyskali Valley 1977 |
Predator of aquatic snails in the water |
[84] |
F270 |
JN860447 |
JN837575 |
JN816333 |
JN837506 |
|
| |
|
|
|
F271 |
JN860448 |
JN837576 |
JN816334 |
JN837507 |
JN816254 |
| |
Dictya stricta Steyskal 1938 |
Predator of aquatic snails in the water |
[84] |
F260 |
JN860449 |
JN837577 |
|
JN837508 |
|
| |
Dictya texensis Curran 1932 |
Predator of aquatic snails in the water |
[84] |
F268 |
JN860450 |
JN837578 |
JN816335 |
JN837509 |
|
| |
Dictyacium |
|
|
|
|
|
|
|
|
| |
Dictyacium firmum Steyskal 1956 |
Unknown |
|
F187 |
JN860451 |
JN837579 |
JN816337 |
JN837510 |
|
| |
|
|
|
F188 |
JN860452 |
JN837580 |
JN816338 |
JN837511 |
|
| |
Elgiva |
|
|
|
|
|
|
|
|
| |
Elgiva connexa Steyskal 1954 |
Predator of aquatic snails in the water |
[86] |
F150 |
AY875153 |
AY875184 |
AY875091 |
AY875122 |
|
| |
|
|
|
F151 |
JN860453 |
JN837581 |
JN816282 |
JN837512 |
|
| |
|
|
|
F152 |
JN860454 |
JN837582 |
JN816283 |
JN837513 |
JN816255 |
| |
Elgiva solicita (Harris 1780) |
Predator of aquatic snails in the water |
[86] |
F5 |
AY875154 |
AY875185 |
AY875092 |
AY875123 |
|
| |
|
|
|
F6 |
JN860455 |
JN837583 |
JN816284 |
JN837514 |
|
| |
Ethiolimnia |
|
|
|
|
|
|
|
|
| |
Ethiolimnia geniculata (Loew 1862) |
Unknown |
|
F255 |
JN860456 |
JN837584 |
JN816339 |
JN837515 |
JN816256 |
| |
Euthycera |
|
|
|
|
|
|
|
|
| |
Euthycera arcuata (Loew 1859) |
Parasitoid of slugs |
[87] |
F222 |
JN860457 |
JN837585 |
JN816340 |
|
JN816257 |
| |
|
|
|
F223 |
JN860458 |
JN837586 |
JN816341 |
JN837516 |
|
| |
|
|
|
F224 |
JN860459 |
JN837587 |
JN816342 |
JN837517 |
|
| |
Hedria |
|
|
|
|
|
|
|
|
| |
Hedria mixta Steyskal 1954 |
Predator of submerged aquatic snails |
[88] |
F168 |
JN860460 |
JN837588 |
JN816285 |
JN837518 |
|
| |
|
|
|
F169 |
AY875155 |
AY875186 |
AY875093 |
AY875124 |
|
| |
Hoplodictya |
|
|
|
|
|
|
|
|
| |
Hoplodictya acuticornis (Wulp 1897) |
Parasitoid of succineid snails |
LV Knutson (pers. comm.) |
F277 |
JN860461 |
JN837589 |
JN816343 |
JN837519 |
JN816258 |
| |
|
|
|
F278 |
JN860462 |
JN837590 |
JN816344 |
JN837520 |
|
| |
Hydromya |
|
|
|
|
|
|
|
|
| |
Hydromya dorsalis (Fabricius 1775) |
Predator of shoreline-stranded aquatics |
[89] |
F249 |
JN860463 |
JN837591 |
JN816345 |
JN837521 |
JN816259 |
| |
Ilione |
|
[90] |
|
|
|
|
|
|
| |
Ilione albiseta (Scopoli 1763) |
Predator of submerged aquatic snails |
[91,92] |
F122 |
JN860464 |
JN837592 |
JN816286 |
|
|
| |
Limnia |
|
|
|
|
|
|
|
|
| |
Limnia boscii Robineau-Desvoidy 1830 |
Parasitoid of succineid snails |
LV Knutson (pers. comm.) |
F120 |
AY875156 |
AY875187 |
AY875094 |
AY875125 |
JN816260 |
| |
|
|
|
F121 |
JN860465 |
JN837593 |
JN816287 |
JN837522 |
|
| |
Limnia ottawensis Melander 1920 |
Unknown |
|
F154 |
AY875157 |
AY875188 |
AY875095 |
AY875126 |
JN816261 |
| |
Limnia sandovalensis Fisher & Orth 1978 |
Unknown |
|
F155 |
AY875158 |
AY875189 |
AY875096 |
AY875127 |
|
| |
|
|
|
F156 |
JN860466 |
JN837594 |
JN816288 |
JN837523 |
|
| |
Pherbecta |
|
|
|
|
|
|
|
|
| |
Pherbecta limenitis Steyskal 1956 |
Unknown |
|
F237 |
JN860467 |
JN837595 |
JN816346 |
JN837524 |
JN816262 |
| |
Pherbina |
|
|
|
|
|
|
|
|
| |
Pherbina coryleti (Scopoli 1763) |
Predator of shoreline-stranded aquatics |
[37] |
F250 |
JN860468 |
JN837596 |
JN816347 |
|
|
| |
Poecilographa |
|
|
|
|
|
|
|
|
| |
Poecilographa decora (Loew 1864) |
Unknown |
|
F212 |
JN860469 |
JN837597 |
JN816348 |
JN837525 |
JN816263 |
| |
|
|
|
F230 |
JN860470 |
JN837598 |
JN816349 |
JN837526 |
|
| |
Psacadina |
|
|
|
|
|
|
|
|
| |
Psacadina zernyi (Mayer 1953) |
Predator of shoreline-stranded aquatics |
[37] |
F251 |
JN860471 |
|
JN816350 |
JN837527 |
JN816264 |
| |
Renocera |
|
|
|
|
|
|
|
|
| |
Renocera amanda (Cresson 1920) |
Parasitoid of fingernail clams below the water's surface |
[93] |
F88 |
AY875159 |
AY875190 |
AY875097 |
AY875128 |
|
| |
Renocera johnsoni (Cresson 1920) |
Predator of aquatic snails in the water |
BA Foote (unpublished) |
F90 |
AY875160 |
AY875191 |
AY875098 |
AY875129 |
|
| |
|
|
|
F92 |
JN860472 |
JN837599 |
JN816289 |
JN837528 |
JN816265 |
| |
Renocera pallida (Fallen 1820) |
Parasitoid of fingernail clams above the water's surface |
[94] |
F193 |
JN860473 |
JN837600 |
JN816351 |
JN837529 |
|
| |
|
|
|
F194 |
JN860474 |
JN837601 |
JN816352 |
JN837530 |
JN816266 |
| |
Sepedon |
|
|
|
|
|
|
|
|
| |
Sepedon armipes Loew 1859 |
Predator of aquatic snails in the water |
[95] |
F28 |
AY875161 |
AY875192 |
AY875099 |
AY875130 |
|
| |
Sepedon fuscipennis Loew 1859 |
Unknown |
|
F116 |
JN860475 |
JN837602 |
JN816360 |
|
|
| |
|
|
|
F117 |
AY875162 |
AY875193 |
AY875100 |
AY875131 |
|
| |
Sepedon praemiosa Giglio-Tos 1893 |
Predator of aquatic snails in the water |
[95] |
F118 |
AY875163 |
AY875194 |
AY875101 |
AY875132 |
JN816267 |
| |
Tetanocera |
|
|
|
|
|
|
|
|
| |
Tetanocera amurensis Hendel 1809 |
Unknown |
|
F198 |
JN860478 |
JN837605 |
JN816290 |
JN837533 |
|
| |
|
|
|
F199 |
JN860479 |
JN837606 |
JN816291 |
JN837534 |
|
| |
|
|
|
F200 |
JN860480 |
JN837607 |
JN816292 |
|
|
| |
Tetanocera annae Steyskal 1938 |
Predator of aquatic snails in the water |
[96] |
F201 |
JN860481 |
JN837608 |
JN816293 |
|
JN816270 |
| |
|
|
|
F202 |
JN860482 |
JN837609 |
JN816294 |
|
|
| |
|
|
|
F229 |
JN860483 |
JN837610 |
JN816319 |
JN837535 |
|
| |
Tetanocera arnaudi Orth & Fisher 1982 |
Unknown |
|
F23 |
JN860484 |
JN837611 |
JN816295 |
JN837536 |
|
| |
|
|
|
F24 |
JN860485 |
JN837612 |
JN816296 |
JN837537 |
|
| |
Tetanocera arrogans Meigen 1830 |
Parasitoid of succineid snails |
[39] |
F93 |
AY875165 |
AY875196 |
AY875103 |
AY875134 |
JN816271 |
| |
Tetanocera bergi Steyskal 1954 |
Predator of aquatic snails in the water |
[73] |
F159 |
JN860486 |
|
JN816297 |
JN837538 |
JN816272 |
| |
Tetanocera clara Loew 1862 |
Parasitoid of slugs |
[97,98] |
F57 |
AY875167 |
AY875198 |
AY875105 |
AY875136 |
JN816273 |
| |
Tetanocera elata (Fabricius 1781) |
Parasitoid of slugs |
[99] |
F245 |
JN860487 |
JN837613 |
JN816298 |
JN837539 |
|
| |
|
|
|
F247 |
JN860488 |
JN837614 |
JN816299 |
JN837540 |
|
| |
Tetanocera ferruginea Fallén 1820 |
Predator of aquatic snails in the water |
[96] |
F34 |
AY875168 |
AY875199 |
AY875106 |
AY875137 |
|
| |
|
|
|
F158 |
AY875166 |
AY875197 |
AY875104 |
AY875135 |
|
| |
Tetanocera freyi Stackelberg 1963 |
Unknown |
|
F203 |
JN860489 |
JN837615 |
JN816300 |
|
JN816274 |
| |
Tetanocera fuscinervis (Zetterstedt 1838) |
Predator of shoreline-stranded aquatics |
[100] |
F53 |
AY875169 |
AY875200 |
AY875107 |
AY875138 |
|
| |
|
|
|
F54 |
JN860490 |
JN837616 |
JN816302 |
JN837541 |
|
| |
|
|
|
F153 |
JN860491 |
JN837617 |
JN816301 |
JN837542 |
|
| |
Tetanocera hyalipennis Roser 1840 |
Predator of shoreline-stranded aquatics |
[39] |
F127 |
JN860492 |
JN837618 |
JN816303 |
|
|
| |
|
|
|
F191 |
JN860493 |
JN837619 |
JN816304 |
JN837543 |
|
| |
|
|
|
F192 |
JN860494 |
JN837620 |
JN816305 |
JN837544 |
|
| |
Tetanocera kerteszi Hendel 1901 |
Predator of terrestrial snails |
LV Knutson (pers. comm.) |
F46 |
AY875170 |
AY875201 |
AY875108 |
AY875139 |
|
| |
|
|
|
F47 |
JN860495 |
JN837621 |
JN816306 |
JN837545 |
|
| |
Tetanocera latifibula Frey 1924 |
Predator of aquatic snails in the water |
[96] |
F144 |
JN860496 |
JN837622 |
JN816357 |
JN837546 |
|
| |
|
|
|
F146 |
JN860497 |
JN837623 |
JN816358 |
JN837547 |
|
| |
|
|
|
F147 |
AY875171 |
AY875202 |
AY875109 |
AY875140 |
|
| |
|
|
|
F149 |
JN860498 |
JN837624 |
JN816359 |
JN837548 |
|
| |
Tetanocera loewi Steyskal 1959 |
Predator of aquatic snails in the water |
[96] |
F189 |
JN860499 |
JN837625 |
JN816307 |
JN837549 |
|
| |
|
|
|
F226 |
JN860500 |
JN837626 |
JN816308 |
JN837550 |
|
| |
Tetanocera melanostigma Steyskal 1959 |
Parasitoid of succineid snails |
[101] |
F2 |
AY875172 |
AY875203 |
AY875110 |
AY875141 |
|
| |
Tetanocera mesopora Steyskal 1959 |
Predator of aquatic snails in the water |
[96] |
F40 |
AY875173 |
AY875204 |
AY875111 |
AY875142 |
|
| |
Tetanocera montana Day 1881 |
Predator of aquatic snails in the water |
[96] |
F142 |
AY875174 |
AY875205 |
AY875112 |
AY875143 |
|
| |
|
|
|
F143 |
JN860501 |
JN837627 |
JN816309 |
JN837551 |
|
| |
|
|
|
F170 |
JN860502 |
JN837628 |
JN816310 |
JN837552 |
|
| |
|
|
|
F171 |
JN860503 |
JN837629 |
JN816311 |
JN837553 |
JN816275 |
| |
Tetanocera obtusifibula Melander 1920 |
Predator of aquatic snails in the water |
[96] |
F275 |
JN860504 |
JN837630 |
JN816353 |
JN837554 |
|
| |
|
|
|
F276 |
JN860505 |
JN837631 |
JN816354 |
JN837555 |
|
| |
Tetanocera oxia Steyskal 1959 |
Parasitoid of succineid snails |
[101] |
F204 |
JN860506 |
JN837632 |
JN816312 |
|
|
| |
Tetanocera phyllophora Melander 1920 |
Predator of terrestrial snails |
[98,102] |
F39 |
AY875175 |
AY875206 |
AY875113 |
AY875144 |
|
| |
Tetanocera plebeja Loew 1862 |
Parasitoid of slugs |
[97,98] |
F1 |
JN860507 |
JN837633 |
JN816314 |
JN837556 |
|
| |
|
|
|
F13 |
AY875176 |
AY875207 |
AY875114 |
AY875145 |
|
| |
|
|
|
F205 |
JN860508 |
JN837634 |
JN816313 |
|
JN816276 |
| |
Tetanocera plumosa Loew 1847 |
Predator of aquatic snails in the water or on damp shorelines |
[73] |
F11 |
AY875177 |
AY875208 |
AY875115 |
AY875146 |
|
| |
|
|
|
F43 |
JN860509 |
JN837635 |
|
JN837557 |
|
| |
Tetanocera robusta Loew 1847 |
Predator of aquatic snails in the water |
[96] |
F10 |
AY875178 |
AY875209 |
AY875116 |
AY875147 |
|
| |
|
|
|
F16 |
JN860510 |
JN837636 |
JN816317 |
JN837558 |
|
| |
|
|
|
F134 |
JN860511 |
JN837637 |
JN816315 |
JN837559 |
JN816277 |
| |
|
|
|
F137 |
JN860512 |
JN837638 |
JN816316 |
JN837560 |
|
| |
Tetanocera rotundicornis Loew 1861 |
Parasitoid of succineid snails |
[101] |
F206 |
JN860513 |
JN837639 |
JN816318 |
JN837561 |
|
| |
Tetanocera silvatica Meigen 1830 |
Predator of shoreline-stranded aquatics |
[100] |
F35 |
JN860515 |
|
JN816321 |
JN837562 |
JN816279 |
| |
|
|
|
F172 |
AY875179 |
AY875210 |
AY875117 |
AY875148 |
JN816278 |
| |
|
|
|
F173 |
JN860514 |
JN837640 |
JN816320 |
JN837563 |
|
| |
Tetanocera soror Melander 1920 |
Predator of aquatic snails in the water |
[103] |
F209 |
JN860516 |
JN837641 |
JN816322 |
|
|
| |
|
|
|
F210 |
JN860517 |
JN837642 |
JN816323 |
|
|
| |
Tetanocera valida Loew 1862 |
Parasitoid of slugs |
[97,98] |
F84 |
AY875180 |
AY875211 |
AY875118 |
AY875149 |
JN816280 |
| |
Tetanocera vicina Macquart 1843 |
Predator of aquatic snails in the water |
[96] |
F94 |
AY875181 |
AY875212 |
AY875119 |
AY875150 |
|
| |
|
|
|
F95 |
JN860518 |
JN837643 |
JN816324 |
JN837564 |
|
| |
|
|
|
F98 |
JN860519 |
JN837644 |
JN816325 |
JN837565 |
|
| |
|
|
|
F99 |
JN860520 |
JN837645 |
JN816326 |
JN837566 |
|
| |
Trypetoptera |
|
|
|
|
|
|
|
|
| |
Trypetoptera canadensis (Macquart 1843) |
Predator of terrestrial snails |
BA Foote (unpublished) |
F164 |
AY875164 |
AY875195 |
AY875102 |
AY875133 |
JN816268 |
|
Trypetoptera punctulata (Scopoli 1763) |
Predator of terrestrial snails |
|
[104] |
F217 |
JN860476 |
JN837603 |
JN816355 |
JN837531 |
JN816269 |
| |
|
|
|
F218 |
JN860477 |
JN837604 |
JN816356 |
JN837532 |
|
| Sequence coverage |
out of 114 OTUs |
out of 65 species |
out of 23 genera |
||||||
| COI |
114: 100% |
65: 100% |
23: 100% |
||||||
| COII |
110: 96.5% |
62: 95.4% |
21: 91.3% |
||||||
| 16S |
111: 97.4% |
63: 96.9% |
22: 95.6% |
||||||
| 28S |
101: 88.6% |
60: 92.3% |
21: 91.3% |
||||||
| EF1α | 34: 29.8% | 33: 50.8% | 19: 82.6% | ||||||
In a previous paper, a DNA sequence-based phylogeny of sciomyzids was used to examine the evolution of larval characters that appeared correlated with larval habitat [30]. Character states in four larval characters were found to be significantly correlated with aquatic to terrestrial transitions in Tetanocera where each larval character changed in the same way as multiple lineages made independent habitat transitions. In the present study, we build on these findings by examining feeding behavior evolution, as feeding behaviors are dependent on both larval morphological adaptations to different environments and specific requirements related to finding and subduing different prey species. Given the diversity of feeding behaviors within Sciomyzidae and Tetanocera, it is important to determine whether there were single or multiple origins of feeding behaviors. Such an analysis would simultaneously show whether there was convergent evolution of larval habitat and the relative frequencies of habitat transitions. Multiple evolutionary hypotheses regarding feeding behaviors and habitat transitions are presented in the literature (e.g., [17,18,20,21,23,27-29]) and all should be considered plausible until rigorously evaluated using modern phylogenetic comparative methods. Therefore, the present study has four specific objectives: (i) construct a robust estimate of phylogeny for Tetanocera and Tetanocerini based on multiple mitochondrial and nuclear genes, (ii) estimate the evolutionary transitions in larval feeding behaviors, habitats and host/prey that have occurred during the evolution of Tetanocerini and Tetanocera, (iii) test prior hypotheses regarding the monophyly of feeding and ecological groupings and (iv) identify the mechanisms underlying habitat and feeding behavior evolution in Tetanocera.
Results
Phylogenetic analyses
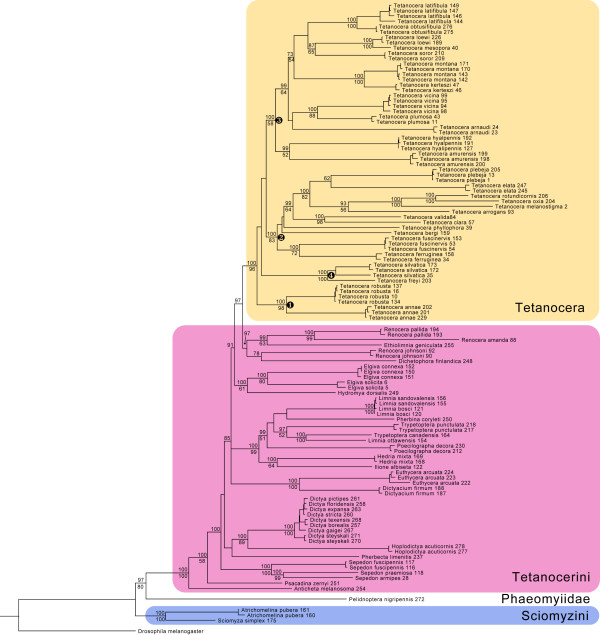
We used Bayesian inference (BI) and maximum likelihood (ML) to analyze a concatenated 5-gene data set. The BI MAP tree with BI posterior probabilities (x100) and ML bootstrap nodal support values is shown in Figure 1. The BI MAP and best ML tree (Additional file 1: Figure S1) were largely congruent (also see Additional file 1: Figure S2, Additional file 1: Figure S3 for BI consensus and ML bootstrap trees, respectively). Both recovered a monophyletic Tetanocerini (BI PP = 1.0; ML bootstrap = 100), a monophyletic Sciomyzini (PP = 1.0; BS = 100), and placed Pelidnoptera, now in Phaeomyiidae but once considered a subfamily of Sciomyzidae (e.g., [31]) as the sister lineage to the Tetanocerini, suggesting its potential status as a tribe within the Sciomyzinae (PP = 0.97; BS = 80; Figure 1). All genera with multiple species are monophyletic except for (1) Limnia, which is rendered polyphyletic by Trypetoptera and Pherbina in the BI MAP tree and by Trypetoptera in the best ML tree, (2) Trypetoptera, rendered polyphyletic by Limnia ottawensis in both trees and (3) Renocera, rendered polyphyletic by Ethiolimnia and Dichetophora in both trees. The polyphyly of these genera are each supported by at least one node with high BI PP and ML BS values (Figure 1).
Figure 1.
Majority rule consensus of 20,000 post burn-in trees from a 160 million generation Bayesian analysis of COI, COI and 16S mtDNA and 28S nuclear DNA from 64 sciomyzid and one phaeomyiid species under a partitioned substitution model. Bayesian posterior probabilities (x100) appear above the nodes and maximum likelihood bootstrap values (200 bootstrap replicates) appear below the nodes. Nodal support values for individuals of the same species were generally high, but were left off due to spatial constraints (as were those for species of Dictya), but appear in the supplemental figures. Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1).
Both BI and ML recovered a monophyletic Tetanocera (PP = 1.0; BS = 96; Figure 1). Within Tetanocera, both trees have T. robusta + T. annae (Figures 1,2,3: clade ➀) as sister to the remaining species. Both analyses recovered Tetanocera clade ➁ (Figures 1,2,3) with identical relationships. This clade includes all five behavioral groups known for the genus (Table 1; Figure 2). The other major Tetanocera clade common to both trees (Figures 1,2,3: clade ➂) contains eight aquatic predators, one shoreline predator, one terrestrial predator and two species with unknown life cycles, with relatively minor differences in species relationships between the BI and ML trees. Finally, both analyses recovered T. silvatica + T. freyi (Figures 1,2,3: clade ➃) as sister species: in the BI MAP tree, clade ➃ is sister to clade ➁ + clade ➂, however in the best ML tree, clade ➃ is sister to clade ➂ (Additional file 1: Figure S1). The BI MAP and best ML trees were not significantly different from one another as judged by ML methods via GARLI and CONSEL (Table 2).
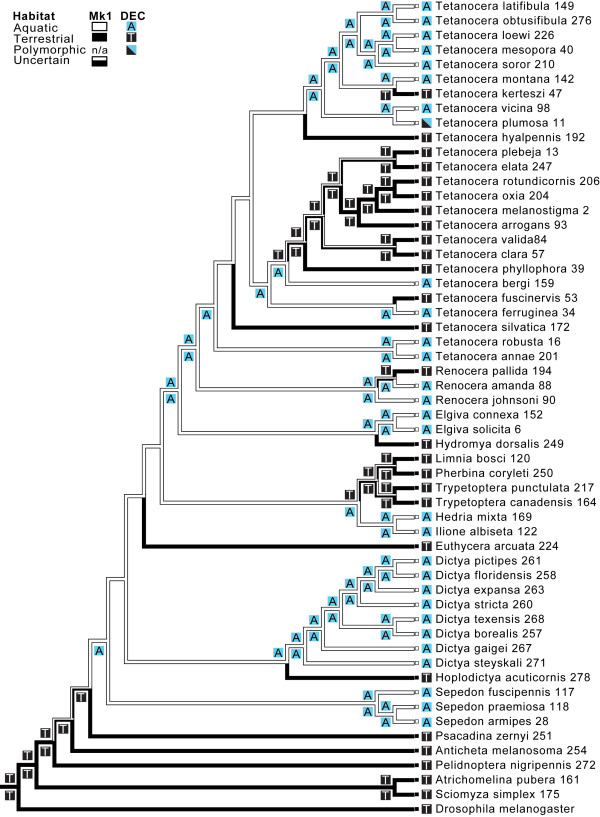
Figure 2.
Maximum likelihood optimization of Knutson and Vala's[17]larval feeding groups on the topology from Figure1(pruned to include only one terminal per species) analyzed with Mesquite using the MK1 model of character evolution. Only character states that are statistically significantly better than the others (ancestral character state estimates with a log likelihood two or more units higher than all others) are shown in the pie charts at the nodes. A solid (one color) node indicates that state is significantly better than all other possible states. Grey indicates unknown character states. Numbers after species names are specimen numbers (Table 1).
Figure 3.
Maximum likelihood optimization of larval habitat on the topology from Figure1(pruned to include only one terminal per species) analyzed with Mesquite using the AsymmMk model of character evolution. Only character states that are statistically significantly better than the others are shown along the branches. A solid (one color) node indicates that state is significantly better than all other possible states. Numbers after species names are specimen numbers (Table 1). Lagrange-estimated ancestral charater states are denoted by blue (aquatic) and black (terrestrial) boxes. Only those nodes with a single state estimated to be significantly better than all other states are plotted. The full Lagrange output is shown in Additional file 1: Figure S5.
Table 2.
Results of the likelihood-based approximately unbiased (AU), Shimodiara-Hasegawa (SH), weighted Kishino-Hasegawa (WKH), and weighted Shimodiara-Hasegawa (WSH) tests calculated using CONSEL
| |
|
|
Test |
|||
|---|---|---|---|---|---|---|
| Constraint | -ln L | Difference | AU | SH | WKH | WSH |
|
Tetanocera feeding group analysis (59-taxon data set) | ||||||
| Unconstrained |
−38932.126 |
(Best) |
|
|
|
|
| Aquatic snail predators1* |
−39212.682 |
280.556 |
p = 4e-06 |
p < 1e-100 |
p < 1e-100 |
p < 1e-100 |
| Aquatic snail predators2* |
−39252.447 |
320.321 |
p = 2e-41 |
p < 1e-100 |
p < 1e-100 |
p < 1e-100 |
| Shoreline snail predators1 |
−38971.538 |
39.412 |
p = 0.001 |
p = 0.002 |
p = 0.002 |
p = 0.002 |
| Shoreline snail predators2* |
−39004.527 |
72.401 |
p = 1e-07 |
p = 0.005 |
p < 1e-100 |
p = 4e-05 |
| Slug parasitoids |
−38941.015 |
8.889 |
p = 0.038 |
p = 0.631 |
p = 0.062 |
p = 0.193 |
| Terrestrial snail predators* |
−39051.229 |
119.103 |
p = 1e-08 |
p < 1e-100 |
p < 1e-100 |
p < 1e-100 |
| Renocerinae monophyly analysis (entire 115-taxon data set) | ||||||
| Unconstrained |
−73022.049 |
(Best) |
|
|
|
|
| Renocerinae |
−73056.937 |
34.89 |
p = 0.055 |
p = 0.059 |
p = 0.059 |
p = 0.059 |
| Comparison of Bayes MAP (Figure 1) and best ML (Additional file 1: Figure S1) trees (entire data set) | ||||||
| ML tree |
−72999.441 |
(Best) |
|
|
|
|
| Bayes MAP tree | −73005.315 | 5.875 | p = 0.377 | p = 0.388 | p = 0.388 | p = 0.388 |
Knutson and Vala [17] feeding group constraints were done with an abbreviated data set containing 59 terminal taxa (all Tetanocera plus 4 outgroups). Trees compared were the best topology from unconstrained analysis versus an analysis where the feeding groups (see Table 1) were constrained to be monophyletic. Tetanocera plumosa, which can either live in the water or on the shoreline was coded both ways (Aquatic2 & Shoreline2 = T. plumosa considered a shoreline snail predator). The monophyly of Anticheta + Renocera, proposed as subfamily Renocerinae by Verbeke [74], was tested by constraining them to be outside of the Tetanocerini and Sciomyzini. The Bayesian MAP tree and ML tree were tested to see if they were significantly different from one another. P-values in bold are significant. Constraints with an asterisk (*) were constrained trees that were significantly worse than the unconstrained tree in all statistical tests.
Behavioral group optimizations
We optimized Knutson and Vala's [17] larval behavioral groups on the BI MAP tree using ML methods in Mesquite (Figure 2; see Additional file 1: Figure S4 for optimization on the best ML tree). From these optimizations, we infer that (i) the evolution of aquatic larvae occurred relatively early in tetanocerine phylogenesis, (ii) from this aquatic ancestor, at least 10 lineages made independent, evolutionary reversals to terrestrial existence and, during the process, made at least 15 feeding behavioral transitions, (iii) the ancestor of Tetanocera was a general predator of non-operculate snails just below the water surface and (iv) a minimum of five Tetanocera lineages made independent, evolutionary reversals to terrestrial existence during which at least seven transitions in feeding behaviors occurred. All of these transitions were judged significant by ML criteria. The optimization of larval habitat (Figure 3) demonstrates an identical aquatic to terrestrial transition pattern (as compared to Figure 2) within the Tetanocerini subsequent to the divergence of Sepedon. Removal of species with unknown life cycles had no significant effects on either optimization.
We also estimated the evolution of habitat changes using the dispersal-extinction-cladogenesis (DEC) model implemented in the program Lagrange [32]. We found the optimal ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions was between 11:1 and 13:1 which was significantly better than the null model with no bias in habitat transition rates (i.e., the global ML estimate was more than two log-likelihood units higher; Additional file 1: Table S1) and congruent with the Mesquite optimization. Therefore, the DEC model-estimated ancestral states plotted on Figure 3 are those with Lagrange set to a 12:1 ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions. This procedure significantly estimated a terrestrial habitat for the sciomyzid ancestral lineage, a single terrestrial-to-aquatic transition and five unambiguous aquatic-to-terrestrial transitions. Furthermore, both the DEC and Mesquite optimizations are congruent and complementary such that only one node, near the base of the tree, does not have a significant habitat estimation (Figure 3). The tetanocerine feeding behavior and habitat transitions based on the Mesquite optimizations in Figures 2 and 3 are better visualized in Figure 4.
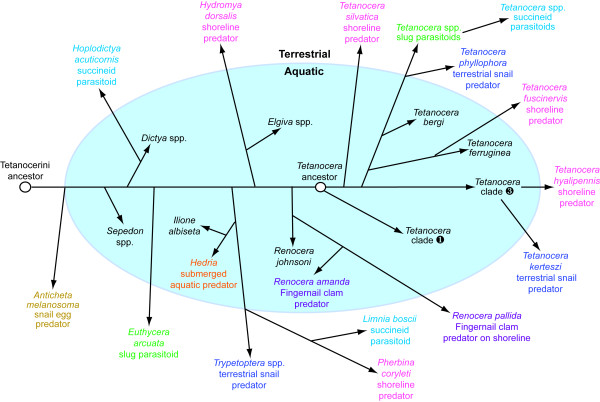
Figure 4.
Diagram showing the evolution of feeding behaviors and habitat changes in the Tetanocerini based on the topology and optimization of Knutson and Vala's[17]behavioral groups in Figure2(unknowns removed). Aquatic lineages in black type are all general predators of aquatic snails. Every line that crosses from blue to white background represents an aquatic-to-terrestrial transition. Branches that split at the aquatic-terrestrial interface indicate uncertainty of ancestral habitat
Constraint analyses
To test hypotheses of multiple independent feeding behavior and habitat transitions, ML analyses were performed in which the monophyly of each polyphyletic Tetanocera behavioral group was constrained (Table 2). In the analyses where (1) aquatic snail predators (two variations), (2) stranded shoreline snail predators (two variations) and (3) terrestrial snail predators were constrained, all topology tests rejected their monophyly as the constrained trees yielded significantly lower log-likelihoods than the unconstrained trees (Table 2). These results support the hypothesis that there were multiple independent transitions from aquatic snail predation in the water to (1) aquatic snail predation on the shoreline and (2) terrestrial snail predation. Constraining the monophyly of slug parasitoid Tetanocera species did not result in a significantly different topology from the unconstrained tree. Finally, constraining Anticheta + Renocera to be monophyletic and further constrained to be a separate lineage outside of the currently recognized tribes produced a tree that was not significantly worse than the unconstrained tree (Table 2).
Discussion
Evolutionary transitions in sciomyzid larvae
Based on life history and larval morphological studies, Knutson & Vala [18] concluded that “terrestrial behavior and morphology in the Tetanocerini are apomorphic features of that tribe.” The present study and others support this conclusion. Wiegmann et al. [33] performed phylogenetic analyses of a comprehensive dipteran data set that yielded a monophyletic Sciomyzoidea that included eight families currently classified in the Sciomyzoidea, the Huttoninidae (elevated to family) and Conopidae, a family not previously included in the Sciomyzoidea (also see [34]). The Sciomyzidae occupies a relatively derived position within the Sciomyzoidea in the Wiegmann et al. [33] phylogeny. The only sciomyzoid taxon known to contain aquatic larvae is the Tetanocerini. Therefore, higher-level studies of Sciomyzoidea [33] support a terrestrial ancestor for Sciomyzidae and their closest relatives. The derived position of Tetanocerini within Sciomyzidae (Figures 12) suggests the freshwater aquatic habit is a unique derived feature of this clade, and terrestrial behavior and morphology in the Tetanocerini are largely derived from aquatic ancestry. Our habitat optimizations (Figure 3) strongly support a terrestrial ancestor for Sciomyzidae and both Mesquite optimizations (Figures 23) strongly support an aquatic ancestor for Tetanocerini minus Anticheta and Psacadina (i.e., the third basal-most Tetanocerini node). These analyses clearly demonstrate a (i) sciomyzid ancestor with terrestrial larva, (ii) derived aquatic habitat for the ancestor of most Tetanocerini and (iii) secondarily derived terrestrial habitat for multiple tetanocerine lineages.
The genus Tetanocera is one of the most remarkable sciomyzid genera with respect to feeding behaviors, microhabitat and host/prey preference as its species are members of five of Knutson and Vala's [17] behavioral groups. The monophyly of Tetanocera is well-supported (BI PP = 1.0; ML bootstrap = 96; Figure 1, Additional file 1: Figure S1), and the ancestor to Tetanocera was strongly suported as being a predator of aquatic snails (Figure 2). From this ancestral condition, there were three independent transitions to shoreline predation on aquatic snails, two independent transitions to terrestrial snail predation, and one transition to slug parasitoidism with one lineage subsequently transitioning to succineid snail parasitoidism (Figures 24). Furthermore, the monophyly of three of these behavioral groups has been rejected (Table 2). In a more general sense, this implies that Tetanocera lineages made between three (Lagrange) and five (Mesquite) independent transitions to terrestrial habitats (Figure 3). These transitions were estimated to be statistically significant by ML (Figures 23, Additional file 1: Figure S4, Additional file 1: Figure S5). In their modification of Steyskal’s [35] morphology-based, Tetanocera species groups, Boyes et al.[36] stated that “the derived, terrestrial modes of [feeding] behavior have clearly arisen several times in different species groups.” Our species-level phylogeny, ML optimizations and topology tests clearly support this conclusion.
Sciomyzids violate trends in habitat transitions
In their study of evolutionary aquatic-terrestrial habitat transitions, Vermeij and Dudley [14] concluded that with the exception of tetrapod vertebrates, aquatic-to-terrestrial habitat transitions are rare as compared to the reverse. However, within Diptera, more than 20 lineages have made such transitions (inferred from Figure 1 of [33]). From the present study, it can be concluded that sciomyzid lineages have made an exceptional number of independent transitions between aquatic and terrestrial habitats. Specifically, they have made at least one transition from terrestrial to aquatic habitats, with Mesquite estimating at least 10 lineages subsequently experiencing evolutionary reversals to terrestrial habitats (Figures 23, Additional file 1: Figure S4). Additionally, the Lagrange analysis demonstrated a significant bias in the transition rate towards aquatic-to-terrestrial transitions with the overall log-likelihood maximizing near a ratio of 12:1 (Additional file 1: Table S2). Furthermore, some species in each of nine tetanocerine genera not included in our analyses (DichetophoraDictyaEulimniaNeolimniaPerilimniaProtodictyaSepedonSepedonellaShannonia) have terrestrial larvae [17], so it is quite likely that there have been additional aquatic-to-terrestrial transitions during the phylogenesis of the Tetanocerini. If we assume that DichetophoraDictyaNeolimniaProtodictya and Sepedon are monophyletic and arose after the early tetanocerine ancestor entered the water (obviously true for three of these genera in Figure 1, Additional file 1: Figure S1), then as many as five additional aquatic-to-terrestrial transitions have occurred, because each of these genera have both aquatic and terrestrial members [17,18]. Given that life cycle, habitat and host/prey information is available for only 240 of 540 sciomyzid species and 41 of the 61 genera [17], the actual number of independent aquatic to terrestrial habitat transitions could easily number in the 20s. Clearly, aquatic-to-terrestrial habitat transitions are strikingly common in the Sciomyzidae.
Possible mechanisms for aquatic to terrestrial transitions
Vermeij and Dudley [14] also concluded that predation intensities are generally lower in freshwater habitats than they are on land, therefore offering less biotic resistance to transitions from terrestrial to freshwater habitats than the reverse. However, our estimated evolutionary transitions within the Tetanocerini show the opposite pattern, with a 10:1 ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions (Figures 234; Additional file 1: Table S2). This raises the question of why sciomyzids are going against the trend observed by Vermeij and Dudley [14]. A portion of the answer likely lies in the larval morphological adaptations necessary for survival in each habitat. Chapman et al.[30] examined changes in four larval characters that were found to be significantly correlated with aquatic-to-terrestrial transitions in Tetanocera. They found that in each independent transition, the larvae of terrestrial lineages experienced reductions or losses in three characters associated with breathing while under water and lost pigmentation (also see [37]). This trend was observed across the Tetanocerini by Vala and Gasc [38], who found a series of reductions in the same breathing-related characters as lineages moved from aquatic to shoreline to drier terrestrial habitats. In order for a terrestrial Tetanocerini lineage to enter the water, it would have to gain those adaptations necessary to respire while mostly submerged. Therefore, the relative ease of losing aquatic adaptations versus the relative difficulty of gaining such adaptations de novo is likely one of the primary reasons that there is a much higher rate of aquatic to terrestrial habitat transitions than the reverse in sciomyzids. This significant reduction in aquatic-to-terrestrial adaptive morphological constraints indicates that tetanocerine phylogenesis likely tracked some ecological pressures (e.g., increased aquatic predation and/or increased terrestrial food availability) more accurately than did more constrained lineages.
In order for a lineage to make a successful transition to a new habitat, the members must be able to compete for and acquire resources in the new habitat [14]. Generally, it seems rather unlikely that a lineage could make multiple parallel transitions into a new habitat as the success of these transitions would typically depend upon a simultaneous adaptation to new physical conditions as well as the utilization of new food resources. However, in the case of the Tetanocerini, the intermediate nature of the damp, shoreline habitat likely played a significant role in facilitating parallel aquatic-to-terrestrial habitat transitions. Our analyses demonstrate that five of the 10 independent transitions to terrestrial habitats were to the shoreline habitat where the prey taxa are the same as their aquatic ancestors (HydromyaRenoceraTetanocera; Figures 24). Both aquatic snails and fingernail clams occur on damp shorelines where they are either periodically stranded by receding or fluctuating waters, or in the case of snails, temporarily foraging out of the water or migrating between aquatic habitats [17]. The availability of the same food resources on the shoreline as in the water likely facilitated stepwise transitions to terrestrial existence in multiple sciomyzid lineages. Like aquatic snails, aquatic sciomyzid larvae can move onto the damp shoreline in search of their prey. However, shoreline-adapted species have lost their adaptations to breathing while under the surface of the water, and will actively swim to shore if placed in water [39]. Once a lineage has adapted to living on damp shorelines (possessing only vestiges of the adaptations to breathing while under water), they may find it more difficult to go back into the water (having to re-express the aquatic-adapted traits) than to move to even drier habitats or switch prey type. Once accustomed to feeding on aquatic snails on terrestrial shorelines, tetanocerine lineages then are pre-adapted to preying on non-aquatic gastropods. Relative to the Tetanocerini, derived terrestrial food items include slugs, succineid (semi-terrestrial) snails and land snails. Given this evolutionary scenario, it is easy to imagine how the ancestors of dry-land terrestrial snail-feeding lineages like TrypetopteraTetanocera phyllophora and T. kerteszi used the shoreline as a stepping-stone environment facilitating a gradual movement to dry-land habitats where they became generalist predators of land snails.
A central question remains as to what selection pressures led multiple lineages of Tetanocerini to transition to terrestrial habitats with some lineages switching to prey other than aquatic pulmonate snails. Chapman et al.[30] speculated that it was a combination of (1) eliminating competition with other aquatic snail predators, (2) compensating for prolonged declines in aquatic snail populations, (3) escaping aquatic insect predators/parasitoids and (4) the reduction and/or loss of suitable aquatic habitats due to the general drying climatic trend that took place between 65 and 5 mya (beginning of Cenozoic era to the end of the Pliocene epoch) that drove these terrestrial transitions. Wiegmann et al.[33], using the penalized likelihood method in r8s [40], estimated that the Sciomyzidae originated ~30 mya (see Fig. S3 in [33]). The oldest known fossil Tetanocerini are preserved in Baltic amber (55–24 mya [41]), and the oldest known fossil Tetanocera (although this generic assignment is questionable [41]) is from the Oligocene epoch (34–24 mya [42,43]). These data place Tetanocerini lineages within the general drying period mentioned above. Therefore, (i) the relative ease of reducing or losing morphological characters (compared to gaining them de novo), (ii) the occurrence of the same prey on damp shorelines as occur in the water and (iii) the general drying trend all likely played key roles resulting in multiple tetanocerine lineages making independent aquatic-to-terrestrial transitions during their phylogenesis.
General implications for evolutionary transitions
The results of this study may have implications for how changes between aquatic and terrestrial habitats have occurred in other animals. Any lineage that (1) occurs in aquatic and terrestrial habitats, (2) respires the same way in aquatic and moist shoreline habitats (e.g., cuticular respiration or open tracheal system) and (3) has the same type of food available in both habitats (e.g., pulmonate snails) could show a similar pattern of multiple independent habitat transitions coincident with changes in behavioral and morphological traits. Borda & Sidall [44] found multiple aquatic-to-terrestrial transitions in arynchobdellid leeches, and Rubinoff [45] found either multiple independent terrestrial-to-aquatic transitions or an evolutionary reversal to terrestrial habitats in one lineage of cosmopterigid moths in Hawaii. Both of these taxa fit the above criteria. Like Sciomyzidae, at least 34 other dipteran families have both aquatic and terrestrial lineages [33] and many of the larger such families have larvae that are, in general, restricted to air-breathing (e.g., Culicidae, Dixidae, Dolichopodidae, Stratiomyidae, Syrphidae, Tipulidae and Tabanidae [13]). Air-breathing insects have open tracheal systems and must establish contact between their spiracles and the atmosphere to respire and must therefore either remain at or come to the surface periodically. Of these families, the Tipulidae (crane flies), unlike many of the families traditionally classified in the suborder Nematocera (primitive flies with long, filamentous antennae) that probably share an aquatic ancestor, may have originated in damp terrestrial, tropical habitats [11]. Wiegmann et al.'s [33] plot of aquatic habitat on their comprehensive dipteran phylogeny indicated that most of the families of suborder Brachycera (derived flies with short antennae) with aquatic lineages were likely of terrestrial origin. Therefore, the findings presented herein should broadly interest anyone studying the evolution of aquatic and terrestrial habitat transitions and associated behavioral and morphological changes in Diptera, a group that includes over 152,000 currently named species [33]. Other lineages that fit the above criteria include oligochaete worms, pulmonate gastropods, decapods, isopods, amphipods, orbatid mites, true bugs in the infraorder Nepomorpha and beetles in the suborder Adephaga, superfamily Byrrhoidea and family Lampyridae. The results of the present study are suggestive that some lineages within these groups will also show multiple convergences on aquatic or terrestrial habits when examined with modern phylogenetic comparative methods.
Conclusions
Phylogenetic analyses of sciomyzid DNA sequences provided strong support that the Sciomyzini, Tetanocerini and Tetanocera are monophyletic (Figure 1). We significantly estimated that (i) the ancestor of the Sciomyzidae was terrestrial (Figures 2, 3), (ii) there was a single terrestrial-to-aquatic transition early in the evolution of the Tetanocerini and, subsequently, (iii) there were at least 10 independent aquatic-to-terrestrial transitions and at least 15 transitions in feeding behaviors (Figures 2, 3, 4, Additional file 1: Figure S2). The 10:1 ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions goes against the general trend observed in animals. We found that the ancestor to Tetanocera was aquatic and five Tetanocera lineages made independent aquatic-to-terrestrial transitions and seven independent transitions in feeding behaviors (Figures 2, 3, Additional file 1: Figure S2). Classifications of sciomyzids into ecological assemblages of species resulted in many non-monophyletic groupings (Figures 2, 3, 4, Additional file 1: Figure S2, Additional file 1: Figure S3) whose monophyly were rejected via phylogenetic constraint analyses (Table 2). Therefore, these findings strongly support our inferences of multiple independent transitions in feeding behaviors, habitats and prey/host usage. The damp shoreline habitat is likely a crucial transitional habitat where tetanocerine lineages that move out of the water to forage can find the same prey taxa as in the water. Once tetanocerine lineages are established on the shoreline, terrestrial molluscan taxa are available as potential food resources. From a morphological standpoint, transitioning from aquatic to terrestrial habitats is easier than the reverse, as adaptations to air-breathing just below the surface of the water are more difficult to gain than to lose. Furthermore, tetanocerine phylogenesis occurred as the Earth was going through a general drying period. These factors likely explain why so many tetanocerine lineages made secondary transitions to terrestrial environments. Finally, the results herein imply that any animal lineage that has aquatic and terrestrial members, respire the same way in both habitats and have the same type of food available in both habitats could show a similar pattern of multiple independent habitat transitions coincident with changes in behavioral and morphological traits.
Materials and methods
Taxon sampling
Phylogenetic analyses were performed on DNA sequences from five genes obtained from 60 Tetanocera specimens (representing 28 species) and 53 individuals representing 21 additional genera within the Sciomyzidae (19 from the Tetanocerini (34 species)), two from the Sciomyzini (2 species) and Pelidnoptera (Phaeomyiidae) which is not currently considered to be a member of the Sciomyzidae but is thought to be its sister taxon ([23,25,26] but see [33]). Therefore, our analyses include 72% of Tetanocera species, 42% of the genera of Tetanocerini, and 15% of the genera of Sciomyzini. Drosophila melanogaster (Drosophilidae) was used as the outgroup in all unconstrained phylogenetic analyses. Table 1 contains a complete listing of the taxa analyzed in this study including GenBank accession numbers and the percentage of OTUs, species and genera sequenced for each gene. For 18 of the 28 Tetanocera species, multiple individuals were available and sequenced for replicate sampling purposes. Of the 29 Tetanocera species with known life cycles, 25 are examined. Of the 41 sciomyzid genera that have behavioral information known for the larvae of at least one species, at least one representative of 17 genera is included. Ten of Knutson and Vala's [17] 15 feeding groups are represented.
Laboratory protocols
Field collections of adult specimens were preserved immediately in 95% ethanol. In the laboratory, specimens were transferred to vials containing 100% hexamethyldisilazane (Polysciences, Inc., Warrington, Pennsylvania, USA) for at least 24 hours, after which the liquid was decanted and the specimens allowed to dry under a fume hood. Prior to preparation for total DNA isolation, the head, legs, wings and abdomen of each specimen were removed from the thorax. Total DNA was isolated from each thorax, and the remaining body parts (which contain the morphological characters necessary for species determination) are stored as vouchers in 95% ethanol at the University of Kentucky. Each specimen and associated DNA extraction was given a unique number. Species identification, collecting locality information and habitat notes were recorded in a database.
Total DNA was isolated from single individuals using Qiagen DNeasy Tissue Kits (QIAGEN Inc., Chatsworth, California, USA) following the manufacturer’s animal tissue protocol. We PCR-amplified fragments of the mitochondrial cytochrome c oxidase subunits I (COI) and II (COII) and 16S rDNA genes, and the nuclear 28S rDNA and elongation factor-1 alpha (EF-1α) genes using the primer pairs listed in Table 3. Each amplicon was purified in NuSieve® GTG® low melting temperature agarose (Lonza, Rockland, Maine, USA) and separated from the agarose with Wizard® PCR preps DNA purification system (Promega Corp., Madison, Wisconsin, USA). PCR reactions (total volume = 50 μL) consisted of 1X Qiagen PCR buffer, 0.2 mM of each dNTP, 0.5 mM of each primer, 1.25 U of Qiagen Taq and 1–5 μL of template DNA. Cycle sequencing protocols followed Folmer et al.[46]; both strands were cycle sequenced using either end-labeled primers (Perkin Elmer AmpliCycle Sequencing Kits; Li-COR sequencer) or labeled dideoxynucleotides (ABI Big-Dye Terminator mix v. 3.0; Applied Biosystems, Foster City, California, USA; ABI sequencer). The separation of cycle sequencing reaction products was done in 3.7% and 5.5% polyacrylamide gels in LI-COR 4200 L-2 and 4200S-2 automated DNA sequencers, respectively, or Applied Biosystems 3730XL or 3730 DNA Analyzers.
Table 3.
Genes / primer information used in this study
| Gene | Primer pair | References | Analyzed fragment size | Notes |
|---|---|---|---|---|
| Mitochondrial loci: | ||||
| 16S |
LR-N-13398 / LR-J-12887 |
[105] |
426 bp |
Primer sequences identical to those of “Locust” |
| COI |
LCO1490 |
[46] |
658 bp |
Together, both COI primer pairs encompass nearly the entire gene |
| |
HCO-700ME |
[106] |
|
|
| |
C1-J-2183 / TL2-N-3014 |
[105] |
813 bp |
|
| COII |
TL2-J-3034 / TK-N-3785 |
[105] |
681 bp |
Amplify all of COII |
| Nuclear loci: | ||||
| 28S |
D1F / D6R |
[107] |
1095 bp |
|
| Ef-1α |
ScioEF1a-F |
Designed herein |
876 bp |
CAYMGDGATTTCATYAARAACATGA |
| ScioEF1a-R | GCRATGTGAGCGGTGTGRCAATCC | |||
Analyzed fragment size is the number of base pairs remaining after primer sequences and regions of ambiguous alignment were removed.
Phylogenetic analyses
Bi-directional sequences were aligned using AlignIR (v. 2.0, LI-COR Biosciences, Inc., Lincoln, Nebraska, USA). Multiple sequence alignments of each gene region were produced with MAFFT [47]. The alignments of the COI, COII and EF-1α sequences contained no indels, however, indels that presented alignment ambiguities were found in the sciomyzid 16S and 28S sequences. The GUIDANCE server [48] was used to assess confidence scores for each column in the MAFFT alignments. Columns with confidence scores < 95% were removed prior to all phylogenetic analyses. The data sets analyzed herein (including program-specific commands) have been deposited on Dryad (http://dx.doi.org/10.5061/dryad.cb098).
Bayesian inference (BI) phylogenetic analyses were conducted on a concatenated (using MacClade v. 4.08 [49]) 4,549-character data set (COI = 1471 nt, COII = 681 nt, 16S = 426 nt, 28S = 1095 nt, EF-1α = 876 nt) with MrBayes (v. 3.1.2 [50,51]). The data set contained 114 terminal taxa for which we generated sequences (including 31 terminals from Chapman et al. [30]), plus one additional terminal (D. melanogaster) whose sequences were obtained from GenBank (Table 1). The data were partitioned by gene region and codon position when appropriate (11 total partitions: three gene regions × three codon positions for the COICOII and EF-1α partitions plus a single partition each for 28S and 16S) and jModeltest (v. 12.9.0 [52]) was used to determine the best-fit model for each partition (Additional file 1: Table S2). To allow each partition to have its own set of parameter estimates, revmattratiostatefreqshape, and pinvar were all unlinked during the analyses. To obtain the most accurate branch length estimates possible, the option prset ratepr = variable (assigns a separate branch length parameter for each partition) was employed as per the recommendations of Marshall et al.[53], who found that BI analyses of partitioned data with a global branch length parameter resulted in significantly longer overall tree length. Four 5-million generation pilot analyses (temp = 0.2, 0.1, 0.02, 0.01) were run to determine the optimal temperature setting to assure an appropriate acceptance rate of swaps between chains [54]. Subsequently, two independent, simultaneous BI searches were run for 160 million generations, saving a tree every 5000 generations, with four search chains each (temp = 0.01). The analysis was terminated ~100 million generations after the average standard deviation of the split frequencies fell below 0.02. The 20,000 post-burn-in trees from each run, determined by examination of the log probability of observing the data by generation plot with Tracer (v. 1.5 [55]), were used to calculate the majority rule consensus tree whose nodal support values were plotted on the BI MAP tree (= maximum a posteriori probability tree).
A maximum likelihood (ML) tree was generated using GARLI (v. 2.0 [56]) using the same partitioning scheme and model assignments as the BI analysis (above) and using the default settings except for the following: searchreps = 5, numberofprecreductions = 20, treerejectionthreshold = 100. The parameter estimates from the search replicate that obtained the tree with the highest log-likelihood value were fixed in a 200-replicate ML bootstrap analysis [57] using default settings.
Character optimizations
The estimation of ancestral feeding groups, based on the BI MAP tree and best ML tree, were carried out using ML methods in Mesquite (v. 2.74 [58]). We followed the behavioral groups of Knutson and Vala [17], which are based on the most recent analysis of sciomyzid life cycles. Our data set included taxa from ten sciomyzid behavioral groups plus Pelidnoptera (Phaeomyiidae) and the outgroup (Drosophila) as 11th and 12th states. The Markov k-state one parameter model (MK1 [59]) was used to infer ancestral character states in the ML optimizations. We also optimized larval habitat (coded as aquatic or terrestrial) for which we utilized the Asymmetrical Markov k-state 2 parameter model (AsymmMK; available only for binary characters [60-62]) which allows forward and backward rates to be different. This model was used because the behavioral group optimization estimated a 10:1 ratio of aquatic-to-terrestrial versus terrestrial-to-aquatic transitions. To make decisions regarding the significance of ancestral character state reconstructions, we followed Pagel [63] (following Edwards [64]) who recommended that ancestral character state estimates with a log-likelihood two or more units lower than the best state estimate (decision threshold [T] set to T = 2) be rejected. Generally viewed as a conservative cutoff, this threshold has been used by numerous recent authors (e.g., [65-69]). The DEC model implemented in the program Lagrange [32] was also used to estimate ancestral habitats using the BI MAP tree. Unlike the Mesquite ML optimization which assumes instantaneous habitat transitions, Lagrange models habitat evolution along branches (i.e., over time), therefore allowing ancestors to occur in two habitats simultaneously. While apparently rare in sciomyzids, we did have one taxon (Tetanocera plumosa; see Hypothesis testing section below) that is known to occur both in aquatic and shoreline habitats. Because the Mesquite optimization suggested a 10:1 ratio of aquatic-terrestrial vs. terrestrial-aquatic transitions, we optimized this parameter in Lagrange and used a 12:1 ratio to infer ancestral habitats.
Hypothesis testing
Because many of Knutson and Vala's [17] behavioral groups within Tetanocera were not monophyletic in the unconstrained analyses, we conducted separate analyses constraining each of these groups to be monophyletic. Each resulting constrained tree was statistically compared (see below) to the unconstrained tree to test whether the monophyly of Knutson and Vala's [17] behavioral groups in Tetanocera could be rejected, thereby adding statistical support to inferences of multiple independent feeding behavior and habitat transitions. One unconstrained and six constrained analyses were done with RAxML (iMAC Pthreads-version [70,71]) using the same partitioning scheme as above under the GTR + G + I model [72]. Twenty replicate searches were done for each analysis (constrained and unconstrained) and the tree with the highest log-likelihood from each was used for topology testing (below). To assure that only topology changes within Tetanocera were the major differences between constrained and unconstrained trees, all but five outgroups were removed, leaving one individual each of D. melanogasterAtrichomelina puberaAnticheta melanosomaHoplodictya acuticornis and Limnia boscii and 54 Tetanocera terminals for which behavioral group is known (see Table 1). The non-Tetanocera taxa remaining in the analyses were chosen because each represents a major lineage in the BI MAP (maximum a-posteriori probability) tree and best ML trees and they had the lowest percentage of missing data. Behavioral groups that were not monophyletic on either the BI MAP or best ML trees were constrained in separate analyses as follows (see Table 1): Aquatic1: all Tetanocera with aquatic larvae including the facultative T. plumosa which can also occur on damp shorelines [73]; Aquatic2: same as Aquatic1 excluding T. plumosa; Shoreline1: all Tetanocera with larvae occurring on damp shorelines and preying on aquatic snails excluding T. plumosa; Shoreline2: same as Shoreline1 plus T. plumosa; Slug: all slug parasitoids; Terrestrials: both species predatory on terrestrial snails.
The full 115-taxon data set was used to test the monophyly of the Renocerinae, proposed by Verbeke [74] to include Renocera + Anticheta, two genera quite distantly separated in the BI and ML analyses presented herein. To constrain Renocera + Anticheta as a lineage outside of the other tribes, the Sciomyzini, Tetanocerini and Renocera + Anticheta were each constrained to be monophyletic with three separate constraint statements. Finally, this data set was also used to evaluate whether the BI MAP tree and the best ML tree were significantly different from one another.
To test for significant differences in topologies between unconstrained and constrained analyses, GARLI (v. 2.0), under the same partitioning scheme and models as the BI analysis, was used to create the site-likelihoods file used as input for the topology-testing program CONSEL (v. 0.1 k [75]). CONSEL was used to do the likelihood-based approximately unbiased test (AU [76]), Shimodaira-Hasegawa test (SH [77]), weighted Kishino-Hasegawa test and weighted Shimodaira-Hasegawa test (WKH and WSH [76]). Results of the KH test [78] were omitted due to its inappropriateness for testing a posteriori significant differences among tree topologies [79].
Competing interests
The authors declare that they have no competing interests.
Author’ contributions
Research was conducted by EGC, under the overall guidance of major professors WRH and BAF at Kent State University and postdoctoral advisor JDH at the University of Kentucky. All advisers were integral to the development, funding and execution of all aspects of research. AAP was involved in data collection of Palaearctic specimens and contributed to the writing of the manuscript.
Supplementary Material
Figure S1. Maximum likelihood tree produced by using the partitioning scheme and model assignments in Additional file 1: Table S2 using the default settings in Garli (v. 2.0) except for the following: searchreps = 5, numberofprecreductions = 20, treerejectionthreshold = 100. Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S2. Bayesian consensus tree produced by using the partitioning scheme and model assignments in Additional file 1: Table S2.Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S3. Maximum likelihood bootstrap tree (200 replicates) produced by using the partitioning scheme and model assignments in Additional file 1: Table S2 using the default settings in Garli (v. 2.0). Parameter estimates from the non-bootstrap search replicate that obtained the tree with the highest log-likelihood value were fixed. Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S4. Maximum likelihood optimization of Knutson and Vala's [17] larval feeding groups on the maximum likelihood topology. Additional file 1: Figure S1; pruned to include only one terminal per species) analyzed with Mesquite using the MK1 model of character evolution. Only character states that are statistically significantly better than the others (ancestral character state estimates with a log likelihood two or more units higher than all others) are shown in the pie charts at the nodes. A solid (one color) node indicates that state is significantly better than all other possible states. Grey indicates unknown character states. Numbers after species names are specimen numbers (Table 1). Table S1. Model log-likelihood scores for variations of the ratio of aquatic-terrestrial:terrestrialaquatic transitions using the DEC model in Lagrange with dispersal and extinction rates for each. Bold font indicates significantly better log-likelihoods (i.e., greater than 2 lnL units) than the null (1:1) model. Red font indicates the parameter setting used in the plot of ancestral character states on Figure 3. Figure S5. Lagrange output with ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions set to 12:1. Only states within 2 log-likelihood units of the best were considered for plotting on Figure 3, and only unambiguous states were plotted. Table S2. Gene information and evolutionary models selected by jModelTest for BI and ML phylogenetic analyses).
Contributor Information
Eric G Chapman, Email: ericgchapman@gmail.com.
Andrey A Przhiboro, Email: littoralia@yahoo.co.uk.
James D Harwood, Email: jharw2@email.uky.edu.
Benjamin A Foote, Email: bfoote@kent.edu.
Walter R Hoeh, Email: whoeh@kent.edu.
Acknowledgments
We thank Lita Greve-Jensen, Jere Kahanpaa, Joe Keiper, Lloyd Knutson, Steve Marshall, Wayne Mathis, Rory McDonnell, Austin Richards, Ladislav Roller, Rudolph Rozkošný and James Zanhiser for donating specimens for this project. This work was supported by NSF grant DEB-0237175 (to WRH), by a Kent State University Graduate Student Senate research grant (to EGC) and by the University of Kentucky Agricultural Experiment Station State project KY008043 (to JDH). We thank Jay Abercrombie, John Leavengood, Lloyd Knutson, and William Murphy and two anonymous reviewers for providing insightful comments on this manuscript. This is publication number 12-08-010 of the University of Kentucky Agricultural Experiment Station.
References
- Stebbins GL, Hill GJC. Did multicellular plants invade the land? Am Nat. 1980;115:343–353. [Google Scholar]
- Shear WA. The early development of terrestrial ecosystems. Nature. 1991;351:283–289. doi: 10.1038/351283a0. [DOI] [Google Scholar]
- Ahlberg PE, Milner AR. The origin and early diversification of tetrapods. Nature. 1994;368:507–514. doi: 10.1038/368507a0. [DOI] [Google Scholar]
- Retallack GJ, Feakes CR. Trace fossil evidence for Late Ordovician animals on land. Science. 1987;235:61–63. doi: 10.1126/science.235.4784.61. [DOI] [PubMed] [Google Scholar]
- Sissom WD. In: The biology of scorpions. Polis GA, editor. Stanford University Press, Stanford; 1990. Systematics, biogeography, and paleontology; pp. 65–160. [Google Scholar]
- Rolfe WDI. In: The terrestrial environment and the origin of land vertebrates. Panchen AL, editor. Academic, London; 1980. Early invertebrate terrestrial faunas; pp. 117–157. [Google Scholar]
- Shear WA, Gensel PG, Jeram AJ. Fossils of large terrestrial arthropods from the Lower Devonian of Canada. Nature. 1996;384:555–557. doi: 10.1038/384555a0. [DOI] [Google Scholar]
- Humphreys GS. In: Advances in Regolith. Roach IC, editor. CRC, LEME; 2003. Evolution of terrestrial burrowing invertebrates; pp. 211–215. [Google Scholar]
- Poinar GO. The evolutionary history of nematodes as revealed in stone, amber and mummies. Brill, Leiden; Boston; 2011. [Nematology Monographs and Perspectives, vol 9] [Google Scholar]
- Labandeira CC, Beall BS, Hueber FS. Early insect diversification: evidence from a Lower Devonian bristletail from Québec. Science. 1988;242:913–916. [Google Scholar]
- Pritchard G, McKee MH, Pike EM, Scrimgeour GJ, Sloty J. Did the first insects live in water or in air? Biol J Linnean Soc. 1993;49:31–44. doi: 10.1111/j.1095-8312.1993.tb00683.x. [DOI] [Google Scholar]
- D’Haese CA. Were the first springtails semi-aquatic? A phylogenetic approach by means of 28s rDNA and optimization alignment. P Roy Soc Lond B Bio. 2002;269:1143–1151. doi: 10.1098/rspb.2002.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt RW, Cummins KW, Berg MB. An Introduction to the Aquatic Insects of North America. Kendall/Hunt. , ; 2008. [Google Scholar]
- Vermeij GJ, Dudley R. Why are there so few evolutionary transitions between aquatic and terrestrial ecosystems? Biol J Linnean Soc. 2000;70:541–554. doi: 10.1111/j.1095-8312.2000.tb00216.x. [DOI] [Google Scholar]
- Chase JM. Are there real differences among aquatic and terrestrial food webs? Tree. 2000;15:408–412. doi: 10.1016/s0169-5347(00)01942-x. [DOI] [PubMed] [Google Scholar]
- Shurin JB, Gruner DS, Hillebrand H. All wet or dried up? Real differences between aquatic and terrestrial food webs. Proc R Soc B. 2006;273:1–9. doi: 10.1098/rspb.2005.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson LV, Vala J-C. Biology of Snail-Killing Sciomyzidae Flies. Cambridge University Press, Cambridge; 2011. [Google Scholar]
- Knutson LV, Vala J-C. An evolutionary scenario of Sciomyzidae and Phaeomyiidae (Diptera) Ann Soc Entomol Fr. 2002;38:145–162. [Google Scholar]
- Coupland JB, Barker GM. In: Natural Enemies of Terrestrial Molluscs. Barker GM, editor. CABI, Oxfordshire; 2004. Diptera as predators and parasitoids of terrestrial gastropods, with emphasis on Phoridae, Calliphoridae, Sarcophagidae, Muscidae and Fanniidae (Diptera, Brachycera, Cyclorrhapha) pp. 85–158. [Google Scholar]
- Ferrar P. A guide to the breeding habits and immature stages of Diptera Cyclorrhapha (part 1: text) E. J. Brill/Scandanavian Science Press, Leiden and Copenhagen; 1987. Sciomyzidae; pp. 815–827. [Google Scholar]
- Berg CO, Knutson LV. Biology and systematics of the Sciomyzidae. Ann Rev Entomol. 1978;23:239–258. doi: 10.1146/annurev.en.23.010178.001323. [DOI] [Google Scholar]
- Greathead DJ. Arthropod natural enemies of bilharzia snails and the possibilities for biological control. Biocontr News Info, CIBC. 1981;2:197–202. [Google Scholar]
- Barker GM, Knutson LV, Vala J-C, Coupland JB, Barnes JK. In: Natural Enemies of Terrestrial Molluscs. Barker GM, editor. CABI, Oxfordshire; 2004. Overview of the biology of marsh flies (Diptera: Sciomyzidae), with special reference to predators and parasitoids of terrestrial gastropods; pp. 159–226. [Google Scholar]
- Wagner R, Bartak M, Borkent A, Courtney G, Goddeeris B, Haenni J-P, Knutson LV, Pont A, Rotheray GE, Rozkošný R, Sinclair B, Woodley N, Zatwarnicki T, Zwick P. Global diversity of dipteran families (Insecta Diptera) in freshwater (excluding Simulidae, Culicidae, Chironomidae, Tipulidae and Tabanidae) Hydrobiologia. 2008;595:489–519. doi: 10.1007/s10750-007-9127-9. [DOI] [Google Scholar]
- Marinoni L, Mathis WN. A cladistic analysis of Sciomyzidae Fallen (Diptera) Proc Entomol Soc Wash. 2000;113:162–209. [Google Scholar]
- Griffiths GCD. The phylogenetic classification of Diptera. Cyclorrhapha with special reference to the structure of the male postabdomen. Junk, The Hague; 1972. [Google Scholar]
- Berg CO, Foote BA, Neff SE. Evolution of predator–prey relationships in snail-killing sciomyzid larvae (Diptera) Bulletin of the American Malacological Union Inc. 1959;25:10–13. [Google Scholar]
- Foote BA, Neff SE, Berg CO. Biology and immature stages of Atrichomelina pubera (Diptera: Sciomyzidae) Ann Entomol Soc Am. 1960;53:192–199. [Google Scholar]
- Steyskal GC. The subfamilies of Sciomyzidae of the world (Diptera: Acalyptratae) Ann Entomol Soc Am. 1965;58:593–594. [Google Scholar]
- Chapman EG, Foote BA, Malukiewicz J, Hoeh WR. Parallel evolution of larval morphology and habitat in the snail-killing fly genus Tetanocera. J Evol Biol. 2006;19:1459–1474. doi: 10.1111/j.1420-9101.2006.01132.x. [DOI] [PubMed] [Google Scholar]
- Rozkošný R. A Review of the Palaearctic Sciomyzidae (Diptera). Folia Facultatis Scientiarum Naturalium Universitatis Purkynianae Brunensis. , ; 1987. [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, Lambkin C, Bertonea MA, Cassel BK, Keith MKM, Heimberg AM, Wheeler BM, Peterson KJ, Pape T, Sinclair BJ, Skevington JH, Blagoderov V, Caravas J, Kutty SN, Schmidt-Ottm U, Kampmeiern GE, Thompson FC, Grimaldi DA, Beckenbach AT, Courtney GW, Friedrichk M, Meier R, Yeates DK. Episodic radiations in the fly tree of life. Proc Nat Acad Sci. 2011; : . doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JF, Skevington JH, Kelso S. Placement of Conopidae (Diptera) within Schizophora based on mtDNA and nrDNA gene regions. Molec Phylogenet Evol. 2010;56:91–103. doi: 10.1016/j.ympev.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Steyskal GC. The American species of the genus Tetanocera Duméril (Diptera) Pap Mich Acad Sci Arts Lett. 1959;44:55–91. [Google Scholar]
- Boyes JW, Knutson LV, van Brinck JM. Further cytotaxonomic studies of Sciomyzidae with description of a new species, Dichetophora boyesi Steyskal (Diptera: Acalyptratae) Genetica. 1972;43:344–365. [Google Scholar]
- Knutson LV, Rozkošný R, Berg CO. Biology and immature stages of Pherbina and Psacadina (Diptera: Sciomyzidae) Acta Scientiarum Naturalium Academiae Scientiarum Bohemicae Brno. 1975;9:1–38. [Google Scholar]
- Vala J-C, Gasc C. Ecological adaptations and morphological variation in the posterior disc of larvae of Sciomyzidae (Diptera) Can J Zool. 1990;68:517–521. doi: 10.1139/z90-076. [DOI] [Google Scholar]
- Knutson LV. Biology and Immature Stages of Snail-killing Flies of Europe Diptera: Sciomyzidae) Cornell University, PhD Thesis; 1963. [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molec Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Hennig W. Die Acalyptratae des baltischen Bernsteins und ihre Bedeutung für die Erforschung der phylogenetischen Entwicklung dieser Dipteren-Gruppe. Stuttg Beitr Naturkd. 1965;145:1–215. [Google Scholar]
- Théobald N. Les insectes fossiles des terrains oligocènes de France. Memoires de la Societe des Sciences de Nancy. 1937;2:1–473. [Google Scholar]
- Förster B. Die Insekten des "Plattigen Steinmergels" von Brunstatt. Abh Geol Spez.-Karte Elsass-Lotheringen. 1891;3:333–594. [Google Scholar]
- Borda E, Siddall ME. Arhynchobdellida (Annelida: Oligochaeta: Hirudinida): phylogenetic relationships and evolution. Molec Phylogenet Evol. 2004;30:213–225. doi: 10.1016/j.ympev.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Rubinoff D. Phylogeography and ecology of an endemic radiation of Hawaiian aquatic case-bearing moths (Hyposmocoma: Cosmopterigidae) Phil Trans R Soc B. 2008;363:3459–3465. doi: 10.1098/rstb.2008.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh WR, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2006;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38:W23–W28. doi: 10.1093/nar/gkq443. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. MacClade: Analysis of Phylogeny and Character Evolution. Sinauer Assoc., Inc., Sunderland; 2000. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molec Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Marshall DC, Simon C, Buckley TR. Accurate branch length estimation in partitioned Bayesian analyses requires accommodation of among-partition rate variation and attention to branch length priors. Syst Biol. 2006;55:992–1003. doi: 10.1080/10635150601087641. [DOI] [PubMed] [Google Scholar]
- Ronquist F, van der Mark P, Huelsenbeck JP. In: The Phylogenetic Handbook: A practical approach to phylogenetic analysis and hypothesis testing. 2. Lemey P, Salemi M, Vandamme A-E, editor. Cambridge University Press, Cambridge; 2009. Bayesian phylogenetic analysis using MrBayes: Practice; pp. 210–266. [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v 1.5. 2009. http://tree.bio.ed.ac.uk/software/tracer/
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin, PhD dissertation; 2006. http://garli.nescent.org. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.74. , ; 2010. http://mesquiteproject.org. [Google Scholar]
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zoologica Scripta. 1997;26:331–348. doi: 10.1111/j.1463-6409.1997.tb00423.x. [DOI] [Google Scholar]
- Mooers AO, Schluter D. Reconstructing ancestor states with maximum likelihood: support for one- and two-rate models. Syst Biol. 1999;48:623–633. doi: 10.1080/106351599260193. [DOI] [Google Scholar]
- Maddison WP. Confounding asymmetries in evolutionary diversification and character change. Evolution. 2006;60:1743–1746. [PubMed] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states on phylogenies. Syst Biol. 1999;48:612–622. doi: 10.1080/106351599260184. [DOI] [Google Scholar]
- Edwards AWF. Likelihood. Cambridge University Press, Cambridge; 1972. [Google Scholar]
- Moczek AP, Cruickshank TE, Shelby A. When ontogeny reveals what phylogeny hides: Gain and loss of horns during development and evolution of horned beetles. Evolution. 2006;60:2329–2341. [PubMed] [Google Scholar]
- Fernandez AA, Morris MR. Sexual selection and trichromatic color vision in primates: Statistical support for the preexisting-bias hypothesis. Am Nat. 2007;170:10–20. doi: 10.1086/518566. [DOI] [PubMed] [Google Scholar]
- Murphy NP, Carey D, Castro LR, Dowton M, Austin AD. Phylogeny of the platygastroid wasps (Hymenoptera) based on sequences from the 18S rRNA, 28S rRNA and cytochrome oxidase I genes: implications for the evolution of the ovipositor system and host relationships. Biol J Linnean Soc. 2007;91:653–669. doi: 10.1111/j.1095-8312.2007.00825.x. [DOI] [Google Scholar]
- Koepfli KP, Deere KA, Slater GJ, Begg C, Begg K, Grassman L, Lucherini M, Veron G, Wayne RK. Multigene phylogeny of the Mustelidae: Resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 2008;6:10. doi: 10.1186/1741-7007-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SC, Chapman EG, Rogers DC, Senyo DM, Hoeh WR. Evolutionary transitions among dioecy, androdioecy and hermaphroditism in limnadiid clam shrimp (Branchiopoda: Spinicaudata) J Evol Biol. 2009;22:1781–1799. doi: 10.1111/j.1420-9101.2009.01813.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Ott M, Zola J, Aluru S, Stamatakis A. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. Proceedings of the 2007 ACM/IEEE conference on supercomputing. 2007. ISBN: 978-1-59593-764-3.
- Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/S0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Foote BA. Biology and immature stages of snail-killing flies belonging to the genus Tetanocera (Diptera: Sciomyzidae). V. Biology of three additional species having larvae that prey on aquatic pulmonate snails. Ann Carnegie Mus. 2011;79:125–130. doi: 10.2992/007.079.0203. [DOI] [Google Scholar]
- Verbeke J, de Witte MGF. Exploration du Parc National Albert. Hayez, Brussels; 1950. Fasc. 66. Sciomyzidae (Diptera Cyclorrhapha) pp. 1–37. 1933–35. [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molec Biol Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order of the Hominoidea. J Molec Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Goldman N, Anderson JP, Rodrigo RG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- Lindsay SL. Food preferences of Drosophila larvae. Am Nat. 1958;92:279–285. doi: 10.1086/282036. [DOI] [Google Scholar]
- Bailey PT. The millipede parasitoid Pelidnoptera nigripennis (F.) (Diptera: Sciomyzidae) for the biological control of the millipede Ommatoiulus moreleti (Lucas) (Diplopoda: Julida: Julidae) in Australia. B Entomol Res. 1989;79:381–391. doi: 10.1017/S0007485300018381. [DOI] [Google Scholar]
- Knutson LV. Life cycles of snail-killing flies: Pherbellia griseicollis, Sciomyza dryomyzina, S. simplex, and S. testacea (Diptera: Sciomyzidae) Entomol Scand. 1987;18:383–391. doi: 10.1163/187631287X00197. [DOI] [Google Scholar]
- Knutson LV, Abercrombie J. Biology of Antichaeta melanosoma (Diptera: Sciomyzidae), with notes on parasitoid Ichneumonidae (Hymenoptera) Proc Entomol Soc Wash. 1977;79:111–125. [Google Scholar]
- Valley K, Berg CO. Biology, immature stages, and new species of snail killing Diptera of the genus Dictya (Sciomyzidae) Search Agriculture, Entomology. 1977;18:1–44. [Google Scholar]
- McLauglin RE, Dame DA. Rearing Dictya floridensis (Diptera: Sciomyzidae) in a continuously producing colony and evaluation of larvae food sources. J Med Entomol. 1989;26:522–527. doi: 10.1093/jmedent/26.6.522. [DOI] [PubMed] [Google Scholar]
- Knutson LV, Berg CO. Biology and immature stages of snail-killing flies: the genus Elgiva (Diptera: Sciomyzidae) Ann Entomol Soc Am. 1964;57:173–192. [Google Scholar]
- Knutson LV. Biology of snail-killing flies in Sweden (Diptera: Sciomyzidae) Entomol Scand. 1970;1:307–314. doi: 10.1163/187631270X00276. [DOI] [Google Scholar]
- Foote BA. Biology of Hedria mixta (Diptera: Sciomyzidae) Ann Entomol Soc Am. 1971;64:931–941. [Google Scholar]
- Knutson LV, Berg CO. Biology and immature stages of a snail-killing fly, Hydromya dorsalis (Fabricius) (Diptera: Sciomyzidae) Proc R Entomol Soc A. 1963;38:45–58. [Google Scholar]
- Hope Cawdery MJ, Lindsay W. Observations on the decline of the snail (Lymnaea truncata, Linn.) and the liver fluke (Fasciola hepatica, Linn.) on reclaimed western blanket peat and its possible relationship to predation by Hydromya dorsalis (Fab.) Proc R Ir Acad Sem Biol Cont. 1977;Seminar on Biological Control:161–169. [Google Scholar]
- Knutson LV, Berg CO. Biology and immature stages of malacophagous Diptera of the genus Knutsonia Verbeke (Sciomyzidae) Institut royal des Sciences naturelles de Belgique, Bulletin. 1967;43:160. [Google Scholar]
- Lindsay W, McDonnell RJ, Williams CD, Knutson L, Gormally MJ. Biology of the snail-killing fly Ilione albiseta (Scopoli, 1763) (Diptera: Sciomyzidae) Studia Dipterol. 2009;16:245–307. [Google Scholar]
- Foote BA. Biology and larval feeding habits of three species of Renocera (Diptera: Sciomyzidae) that prey on fingernail clams (Mollusca: Sphaeriidae) Ann Entomol Soc Am. 1976;69:121–133. [Google Scholar]
- Horsáková J. Biology and immature stages of the clam-killing fly, Renocera pallida (Diptera: Sciomyzidae) Eur J Entomol. 2003;100:143–151. [Google Scholar]
- Neff SE, Berg CO. Biology and immature stages of malacophagous Diptera of the genus Sepedon (Sciomyzidae) Virg Agric Exp Sta Bull. 1966;566:1–113. [Google Scholar]
- Foote BA. Biology and immature stages of snail-killing flies belonging to the genus Tetanocera (Insecta: Diptera: Sciomyzidae) 3. Life histories of the predators of aquatic snails. Ann Carnegie Mus. 1999;68:151–174. [Google Scholar]
- Trelka DG, Foote BA. Biology of slug-killing Tetanocera (Diptera: Sciomyzidae) Ann Entomol Soc Am. 1970;63:877–895. [Google Scholar]
- Foote BA. Biology and immature stages of snail-killing flies belonging to the genus Tetanocera (Diptera: Sciomyzidae). IV. Life histories of predators of land snails and slugs. Ann Carnegie Mus. 2008;77:301–312. doi: 10.2992/0097-4463-77.2.301. [DOI] [Google Scholar]
- Knutson LV, Stephenson JW, Berg CO. Biology of a slug-killing fly, Tetanocera elata (Diptera: Sciomyzidae) Proceedings of the Malacological society of London. 1965;36:213–220. [Google Scholar]
- Foote BA. Biology and immature stages of snail-killing flies belonging to the genus Tetanocera (Insecta: Diptera: Sciomyzidae). 1. Introduction and life histories of predators of shoreline snails. Ann Carnegie Mus. 1996a;65:1–12. [Google Scholar]
- Foote BA. Biology and immature stages of snail-killing flies belonging to the genus Tetanocera (Insecta: Diptera: Sciomyzidae). 2. Life histories of predators of snails of the family Succineidae. Ann Carnegie Mus. 1996b;65:153–166. [Google Scholar]
- Bhatia ML, Keilinm D. On a new case of parasitism of snail (Vertigo genesii Gredl.) by a dipterous larva. Parasitology. 1937;29:399–408. doi: 10.1017/S0031182000024902. [DOI] [Google Scholar]
- Foote BA. Biology and immature stages of the snail-killing flies belonging to the genus Tetanocera (Diptera: Sciomyzidae) Cornell University, PhD Thesis; 1961. [Google Scholar]
- Vala J-C. Description des stades larvaires et donnees sur la biologie de Trypetoptera punctulata (Diptera, Sciomyzidae) Ann Soc Entomol France. 1986;22:67–77. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- Breton S, Burger G, Stewart DT, Blier PU. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.) Genetics. 2006;172:1107–1119. doi: 10.1534/genetics.105.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-K, O’Foighil D. Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Mol Phylogenet Evol. 2000;14:75–88. doi: 10.1006/mpev.1999.0691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum likelihood tree produced by using the partitioning scheme and model assignments in Additional file 1: Table S2 using the default settings in Garli (v. 2.0) except for the following: searchreps = 5, numberofprecreductions = 20, treerejectionthreshold = 100. Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S2. Bayesian consensus tree produced by using the partitioning scheme and model assignments in Additional file 1: Table S2.Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S3. Maximum likelihood bootstrap tree (200 replicates) produced by using the partitioning scheme and model assignments in Additional file 1: Table S2 using the default settings in Garli (v. 2.0). Parameter estimates from the non-bootstrap search replicate that obtained the tree with the highest log-likelihood value were fixed. Drosophila melanogaster sequences were used to root the analysis. Numbers after species names are specimen numbers (Table 1). Figure S4. Maximum likelihood optimization of Knutson and Vala's [17] larval feeding groups on the maximum likelihood topology. Additional file 1: Figure S1; pruned to include only one terminal per species) analyzed with Mesquite using the MK1 model of character evolution. Only character states that are statistically significantly better than the others (ancestral character state estimates with a log likelihood two or more units higher than all others) are shown in the pie charts at the nodes. A solid (one color) node indicates that state is significantly better than all other possible states. Grey indicates unknown character states. Numbers after species names are specimen numbers (Table 1). Table S1. Model log-likelihood scores for variations of the ratio of aquatic-terrestrial:terrestrialaquatic transitions using the DEC model in Lagrange with dispersal and extinction rates for each. Bold font indicates significantly better log-likelihoods (i.e., greater than 2 lnL units) than the null (1:1) model. Red font indicates the parameter setting used in the plot of ancestral character states on Figure 3. Figure S5. Lagrange output with ratio of aquatic-to-terrestrial vs. terrestrial-to-aquatic transitions set to 12:1. Only states within 2 log-likelihood units of the best were considered for plotting on Figure 3, and only unambiguous states were plotted. Table S2. Gene information and evolutionary models selected by jModelTest for BI and ML phylogenetic analyses).