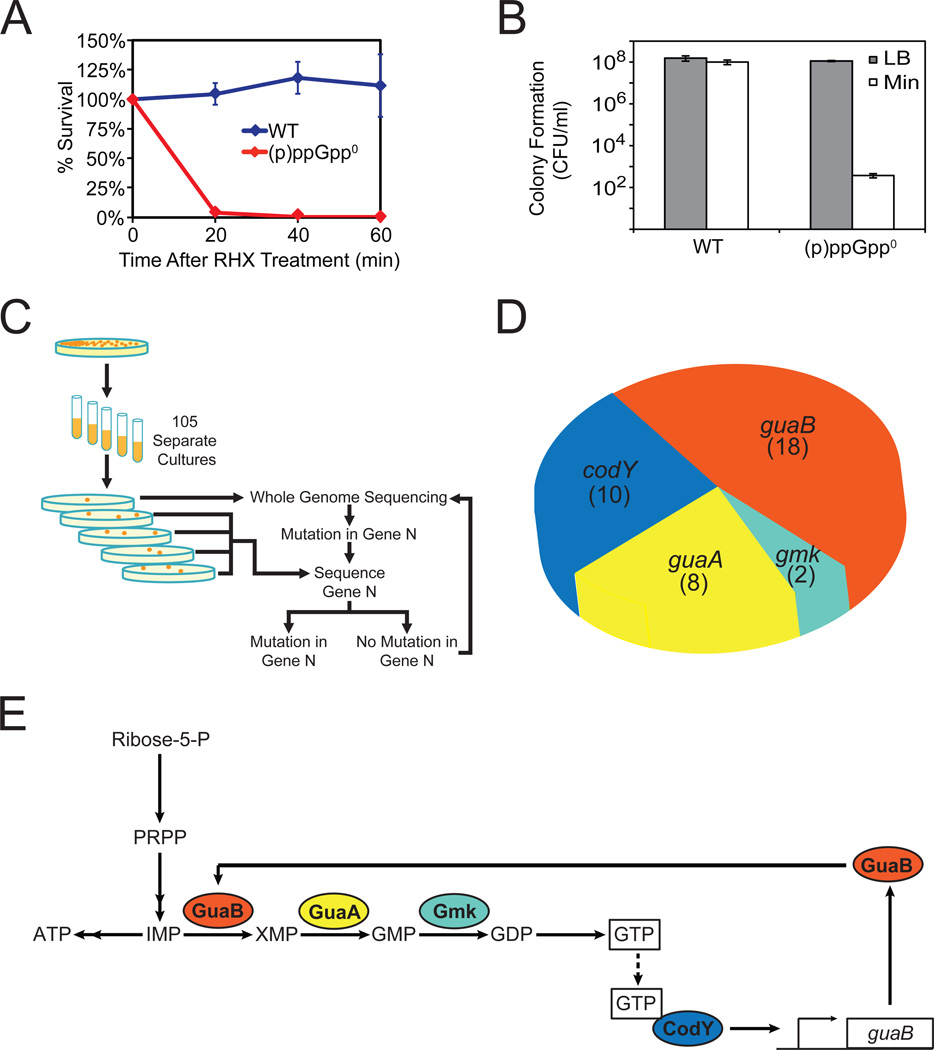
Figure 4. (p)ppGpp0 Phenotype and a Genetic Selection for Suppressor Mutations.
(A) (p)ppGpp0 cells do not survive sudden starvation. WT and (p)ppGpp0 cells were treated with 0.5 mg/ml RHX for the indicated time and plated on LB. Percent survival was calculated by counting the number of cells able to form colonies the next day, normalized to T=0.
(B) (p)ppGpp0 cells fail to form colonies on minimal medium plates. WT and (p)ppGpp0 cells were plated on LB and minimal medium plates, and colonies were counted the next day. CFU/ml: colony forming unit (CFU) per ml of culture, normalized by OD600.
(C) Schematic of genetic selection for (p)ppGpp0 suppressor mutants that can form colonies on minimal medium. Suppressors were selected on minimal medium plates. Whole genome sequencing was conducted on one suppressor to identify a mutation in one gene, followed by targeted sequencing of this gene in all other suppressors. An additional suppressor was subsequently identified by whole-genome sequencing. This process was repeated iteratively.
(D) Distributions of (p)ppGpp0 suppressor mutations in the indicated genes. The numbers indicate independently isolated suppressors with mutations in each gene. Details are listed in Table S5.
(E) Schematic of the pathways affected by the (p)ppGpp0 suppressor mutations. GuaB, GuaA, and Gmk are enzymes in the GTP biosynthesis pathway. CodY is a pleiotropic transcription factor that is activated by GTP and activates transcription of guaB. The colors of the gene products match that of the genes in Figure 4D.