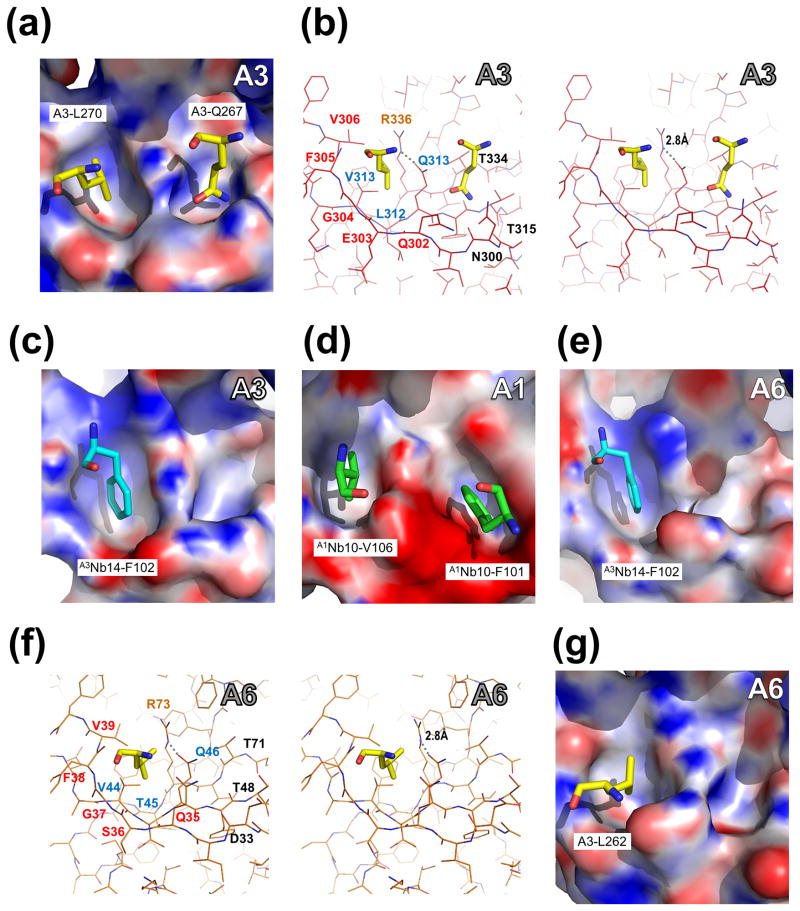
Fig. 4. Pockets of A3OB, A6OB and A1OB with interacting residues in four different crystal structures.
Pocket 1 (left in each panel) and Pocket 2 (right) are defined in Fig. 2(b). The pockets of A3OB, A1OB and A6OB are shown with their electrostatic surface charge. The interacting residues, from either another OB-fold or from a nanobody (Nb), occupying the OB-fold pockets are labeled. The electrostatic potential surface of each OB-fold domain was calculated using APBS (Baker et al., 2001). Regions with potentials above +25 kbTec−1 and below −25 kbTec−1 are shown in blue and red, respectively.
(a – b) Amino acids occupying A3OB Pockets 1 and 2. Pocket 1 and Pocket 2 of A3OB in the current A3OB-9aa-A6 structure occupied by, respectively, A3-L270 and A3-Q267 from another A3OB-9aa-A6 molecule in the crystal, shown in both surface and stereo views. The orientation in panel (b) is the same as in panel (a). The dashed line indicates a hydrogen bond between R336 and Q313. Motif-I, Motif-II and the invariant arginine (R) residues are labeled in red, blue and orange, respectively.
(c) A3Nb14-F102 in A3OB Pocket 1. The A3OB Pocket 1 of the A3OB•A6•(A3Nb14)2 structure (PDB-ID: 3STB; (Park et al., 2012a)) bound with nanobody residue F102 shown in surface view.
(d) A1Nb10-V106 and A1Nb10-F101 in A1OB Pockets 1 and 2. The A1OB Pockets 1 and 2 of the A1OB•A1Nb10 structure (PDB-ID: 4DKA; (Park et al., 2012b)) bound with nanobody residues V106 and F101, respectively, shown in surface view. The corresponding stereo view of these A1 pockets with interacting nanobody residues is shown in Supplementary Fig. 11.
(e) A3Nb14-F102 in A6OB Pocket 1. The A6OB Pocket 1 of the A3OB•A6•(A3Nb14)2 structure (PDB-ID: 3STB; (Park et al., 2012a)) bound with nanobody residue F102 is shown in surface view.
(f – g) A3-L262 in A6OB Pocket 1. The A6 Pocket 1 of the current A3OB-9aa-A6 structure bound with A3-L262 shown in both surface and stereo views. The stereo view in panel (f) has the same orientation as panel (g). The dashed line indicates a hydrogen bond between R73 and Q46. Motif-I, Motif-II and invariant arginine (R) residues are labeled in red, blue and orange, respectively.