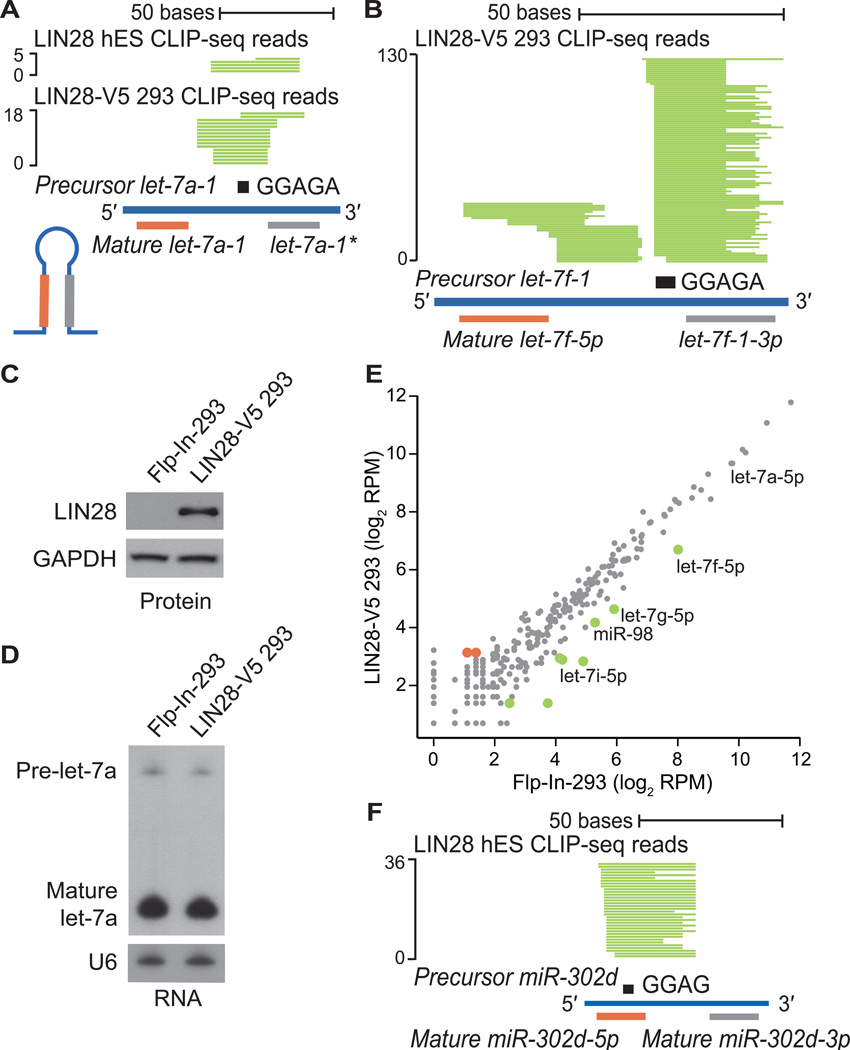
Figure 2. CLIP-seq defines LIN28 binding sites within miRNA precursors.
(A, B) Individual LIN28 CLIP-seq reads (in green) aligned to (A) precursor miRNA let-7a-1 and (B) precursor miRNA let-7f-1, with the mature miRNA boundaries depicted below. The sequence GGAGA in the hairpin loop is depicted as a black rectangle. The scale to the left indicates the number of aligned reads. (C) Western blot analysis of LIN28 protein levels in control Flp-In-293 and LIN28-V5 293 cells. GAPDH serves as a loading control. (D) Northern blot analysis of the human let-7a miRNA in control Flp-In-293 and LIN28-V5 293 cells. The U6 snRNA serves as a loading control. (E) Scatter plot comparing the log2 RPM (reads per million mapped) for expressed mature miRNAs in control Flp-In-293 and LIN28-V5 293 cells (gray), showing significantly upregulated (red) and downregulated (green) miRNAs. (F) LIN28 CLIP-seq reads (in green) aligned to precursor mir-302d, centered on the motif GGAG (black rectangle).