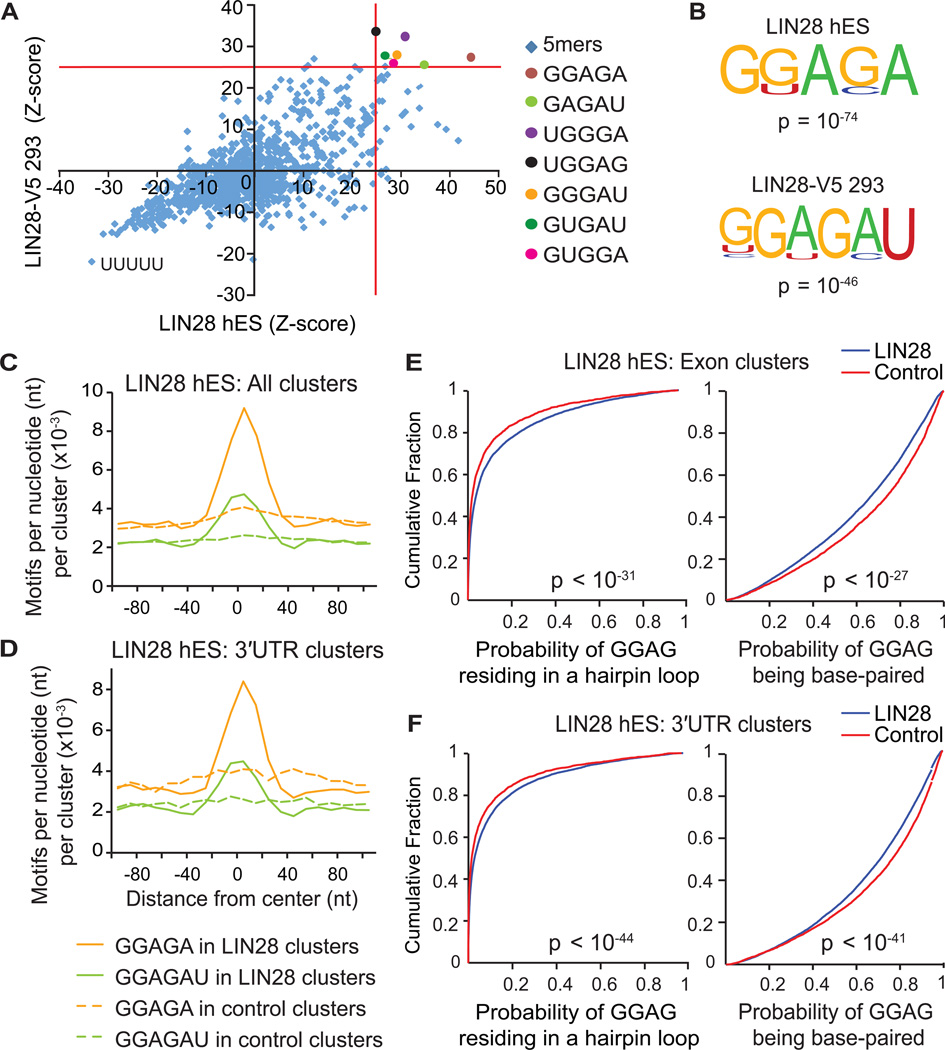
Figure 3. LIN28 binds GGAGA(U) motifs in mRNA sequences within hairpin loop structures.
(A) Scatter plot comparing pentamer Z-scores in hES and LIN28-V5 293 cells. Pentamers overrepresented (p < 10−4) in both cell-types are highlighted by colored circles, and defined on the right. (B) Consensus motifs within LIN28 clusters identified by the HOMER algorithm (Heinz et al., 2010) in hES and LIN28-V5 293 cells with corresponding p-values shown below the motif. (C and D) The positional frequency of consensus motifs GGAGA and GGAGAU relative to the center of (C) all LIN28 hES clusters and (D) clusters only in 3′UTRs. Dashed lines correspond to the positional frequency of these motifs within randomly distributed control clusters from the same type of genic region. (E and F) Cumulative distribution plots display the probability that each nucleotide of a GGAG sequence found within LIN28 hES clusters (blue) or control clusters (red) resides in a predicted hairpin loop (left panel) or base-paired region (right panels) of mRNA; (E) considering clusters only in exons, or (F) clusters only in 3′UTRs (p-values calculated by two-sample Kolgomorov-Smirnov test).