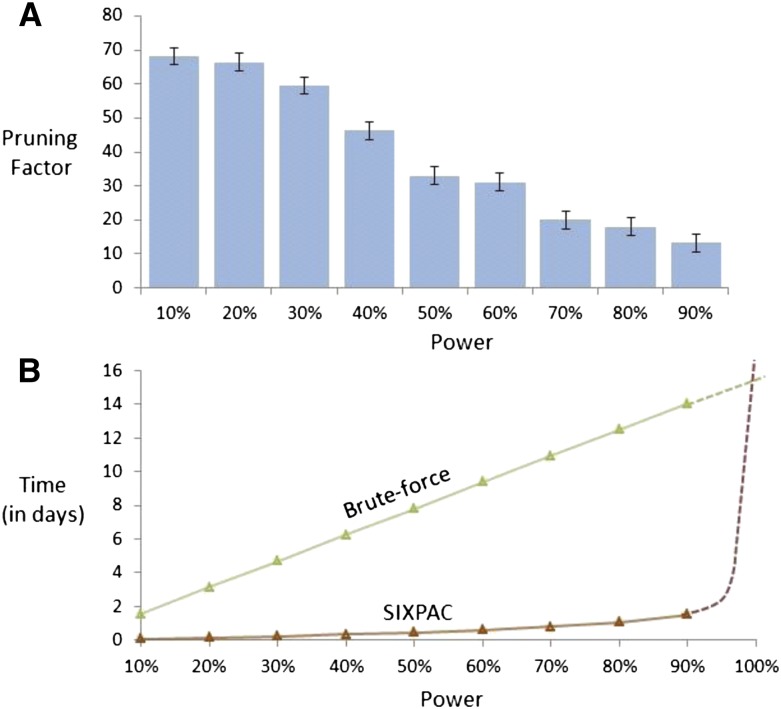
Figure 2.
Computational efficiency. Our implementation of the two-stage PAC-testing framework (SIXPAC, orange line) was benchmarked on the cleaned WTCCC bipolar disorder data set (approximately 2K cases, 3K controls, 450K SNPs, four genetic models tested per distal SNP pair, 400 billion pairwise tests genome-wide). (A) The factor reduction in the universe of SNP pairs achieved by stage 1, for each power setting. Note that unlike brute force, this does not mean down-sampling the universe of SNP pairs, but rather involves reducing the probability of identifying any one of them. For example, a brute-force method would presumably test 40 billion pairs (and ignore the remaining 360 billion) to achieve 10% power on this data set. However, PAC testing scans all 400 billion pairs, but simply reduces the probability of finding the significant interactions among them to 10%. This results in shortlisting ∼68× fewer combinations through stage 1. (B) The efficiency of our software implementation of this method. We compare the performance of SIXPAC against the time taken by a brute-force approach of applying the LD-contrast test directly to all pairs (green line). All tests were benchmarked on a common desktop computer configuration (Intel i7 quad-core processor, 2.67 GHz with 8 GB RAM). The last data point shows the 90% power benchmarks, followed by dotted lines that illustrate how these estimates may continue as we approach 100% power. SIXPAC, like any randomization algorithm, will require infinite compute time to achieve 100% power but can approach very close at a small fraction of the brute-force cost. Lastly, we note that these measurements only reflect the performance of our Java program rather than what might be feasible with a different implementation of the algorithm.