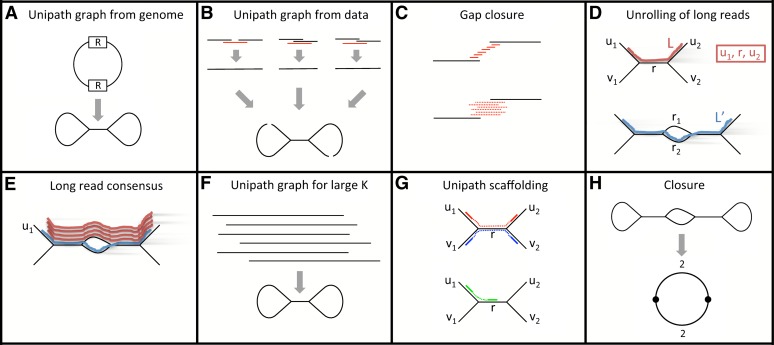
Figure 1.
Diagram of assembly method. (A) The ideal unipath graph depends on the genome and a constant K, the ‘minimum overlap.’ Perfect repeat copies of size K are ‘glued together.’ In the figure, this happens to two copies of a repeat R. (Unipath graphs are actually directed, and both strands of the genome must be accounted for, but we elide these points to facilitate exposition.) (B) As in main text Step I.1, starting from fragment read pairs (data type A), we construct an approximation to the ideal unipath graph. First, individual fragment read pairs are ‘closed’ by recruiting a third read (red; from some other pair). Then the resulting ‘super-reads’ are glued together along perfect repeats of size ≥K. We use K = 96, about half the fragment size. Primarily because of bias introduced by amplification in the sample preparation process, there are gaps in the resulting graph. (C) Gaps in the initial unipath graph are closed either using (top) high-quality bits of jumping reads (data type C, main text Step I.2) or (bottom) lower-quality long reads (data type B, main text Step I.3). (D) Long reads are unrolled along unipath graph as in main text Step II.1. (Top) Long read L is correctly represented as (u1,r,u2). (Bottom) The region contains highly similar unipaths r1 and r2 (perhaps differing by only a single indel base). Long read L′ incorrectly passes through r2 rather than r1, perhaps because it has an error at the same place where r1 and r2 differ. (E) Long read consensus (main text Step II.2). The long read (blue) traverses an incorrect path through the lower part of the middle bubble, whereas several reads (red) traverse the correct upper path, suggesting that a simple voting scheme might work. However, all these reads start at a unipath u1 that is unique in the genome, and it is very challenging to devise heuristics that work well for reads that are not anchored at a unique sequence. (F) Consensus long reads from across the genome are now used to create a unipath graph using K = 640, about half the long read length. Still repeats longer than this K cause the genome to be ‘glued’ together. (G) Unipath scaffolding (main text Step III.2). Jumping pairs are now used to connect unipaths, e.g., u1–u2 and v1–v2 (top), but links to repeats, e.g., u1 to r (bottom) are avoided where possible. (H) Closure (main text Step III.3). (Top) Circular genome whose assembly might be resolved except for a ‘bubble’ in a repeat region (perhaps with branches differing only by a single base). (Bottom) Representation of genome in which vertices represent unambiguous sequence (in this case, nearly all of the genome), and edges represent ambiguous sequences (in this case, two sequences in each of two cases). These edges would correspond to the short unresolved part of the repeat.