SUMMARY
A synthetic genetic system, based on cross-replicating RNA enzymes, provides a means to evaluate alternative genetic codes that relate heritable information to corresponding molecular function. A novel implementation of encoded combinatorial chemistry was used to construct complex populations of cross-replicating RNA enzymes in accordance with a user-specified code that relates genotype and phenotype on a molecule-by-molecule basis. The replicating enzymes were made to undergo self-sustained Darwinian evolution, resulting in the emergence of the most advantageous variants. These included both highly active enzymes that sustained the population as a whole and poorly active enzymes that survived as parasites of the active molecules. This evolutionary outcome was a consequence of the information capacity and fidelity of the genetic code, suggesting how these parameters should be adjusted to implement codes tailored to particular applications.
INTRODUCTION
Genetic systems entail a heritable molecular genotype that encodes a corresponding molecular phenotype. In biology, the genotype resides within the sequence of DNA and the phenotype is expressed as a corresponding set of RNAs and proteins. The vast potential diversity of genetic systems, coupled with the power of natural selection, have led chemists to devise analogous systems for harnessing chemical diversity. The aim has been to merge combinatorial chemistry with either high-throughput screening or selection to discover molecules with desired properties. An especially powerful approach that has been inspired by genetic systems is encoded combinatorial chemistry (Brenner and Lerner, 1992; Gartner and Liu, 2001; Halpin et al., 2004; Melkko et al., 2004; Clark et al., 2009), which links an amplifiable genotype (usually DNA) to corresponding small organic molecules or macromolecules. The linkage must be achieved on a molecule-by-molecule basis, which requires either the separate synthesis of each genotype-phenotype combination or the implementation of a parallel synthesis strategy.
A method that is widely used for assembling combinatorial libraries is split-and-pool synthesis (Furka et al., 1991; Houghten et al., 1991; Thompson and Ellman, 1996). This type of synthesis begins with a common starting material, usually attached to a solid support, which is divided into several equal portions that are reacted separately, each with a different reagent. Then the separate portions are pooled, mixed, and split again. The process is repeated to generate a library of compounds of the desired complexity. With encoded combinatorial chemistry, both the genetic component and the corresponding functional component are added during each split, usually by building off two orthogonal handles that are attached to a common solid support. In the end, the chemical history of each assembled molecular phenotype is recorded in its corresponding amplifiable genotype.
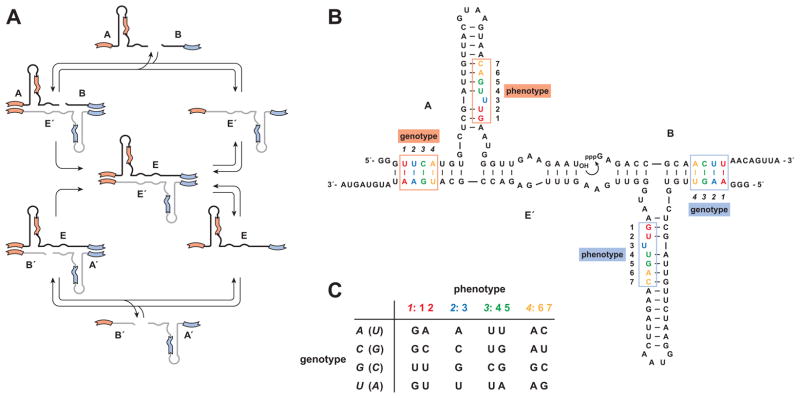
Outside of biology there is currently only one example of a genetic system that can undergo Darwinian evolution in a self-sustained manner (Lincoln and Joyce, 2009). This synthetic genetic system is based on a RNA enzyme with RNA ligase activity (Rogers and Joyce, 2001). The enzyme is configured as a cross-replicating pair (Paul and Joyce, 2002; Kim and Joyce, 2004), whereby two enzymes catalyze each other’s synthesis by joining together component oligonucleotides, resulting in their mutual exponential amplification (Figure 1A). Specifically, a plus-strand RNA enzyme (E) joins two substrates (A′ and B′) to form a minus-strand enzyme (E′), which in turn joins two different substrates (A and B) to form a new plus-stranded enzyme (E). There are two regions of Watson-Crick pairing (6–7 nucleotides each) between the enzyme and substrates that function as genetic loci, enabling the transfer of information from a parent enzyme to its progeny (Figure 1B). Because the sequences of these two regions can vary without markedly affecting replication efficiency, it is possible to construct a population of many different cross-replicating enzymes, each with a different genetic sequence, and allow them to compete for a common set of substrates (Lincoln and Joyce, 2009).
Figure 1.
Cross-replicating RNA enzymes. (A) Cross-replication cycle involving two enzymes, E (black) and E′ (grey), that catalyze each other’s synthesis by joining two component substrates (A′ and B′ to form E′; A and B to form E). (B) Sequence and secondary structure of the wild-type enzyme and its substrates (E′, A, and B are shown). Curved arrow indicates the site of ligation. Boxes indicate nucleotides that can vary within the genotype region (positions 1–4, italics) and corresponding phenotype region (positions 1–7, roman) of the molecule. The A•B′ and A′•B alleles are highlighted in orange and blue, respectively. (C) Genetic code that relates genotype and phenotype positions within A and A′. There is a different code for each of the four genotype positions. The same phenotype nucleotides are encoded by complementary genotype nucleotides in A and A′ (the latter shown in parentheses). (See also Table S1)
As with natural genetic systems, a population of cross-replicating enzymes can be designed such that each distinct genotype encodes a corresponding phenotype, the latter represented as a particular sequence within the functional domain of the enzyme. A genetic code is devised to relate the sequences of the genotype and phenotype regions on a molecule-by-molecule basis. Then, as with encoded combinatorial chemistry, one can carry out repeated rounds of selective amplification to discover functional molecules with desired properties. The set of substrate sequences is fixed, but the utilization of particular substrates is a heritable trait that determines the reproductive fitness of the corresponding enzymes. In addition, a method has been devised to construct an enriched set of substrates derived from the selected enzymes (Lincoln and Joyce, 2009), although that method requires manipulation outside the synthetic genetic system.
The capacity of a population of cross-replicating enzymes to evolve novel function depends on the complexity of the population that can be constructed and the ability to enrich molecules with the desired properties. The initial report of the synthetic evolving system involved only 12 different “alleles” at each of the two genetic loci (Lincoln and Joyce, 2009). Recombination could occur between the two loci, allowing for a combinatorial complexity of 144 different variants. Even this low-complexity system required the synthesis of 72 different components (12 each of the various A, A′, B, B′, E, and E′ molecules). Each of these molecules was synthesized and purified individually, something that would be impractical for a population of thousands of different cross-replicating enzymes. The present study describes a combinatorial method for synthesizing complex libraries of cross-replicating enzymes and their corresponding substrates, enabling one to implement a user-specified genetic code of arbitrary design.
A novel split-and-pool technique was devised that links two regions of the same RNA molecule, one serving as an amplifiable genotype and the other providing a corresponding region of encoded phenotype. The technique involves parallel synthesis of two different arms of a common DNA molecule, which ultimately are ligated to provide a template for PCR amplification and in vitro transcription of the corresponding RNAs. This approach was used to prepare populations of either 4,096 or 65,536 different cross-replicating RNA enzymes and their respective substrates. The syntheses proceeded in accordance with a genetic code that sought to maximize the information capacity at each genetic locus.
The population of 4,096 enzymes was made to undergo self-sustained evolution, involving many successive rounds of selective amplification. Individuals were cloned from the evolved population, sequenced, and analyzed. This analysis revealed that the population was dominated by two types of replicating molecules: those with high catalytic activity that sustained the entire population, and those with poor activity that survived as parasites of the active molecules. The study also revealed advantageous properties of a genetic code that should be considered in the judicious design of synthetic genetic systems.
RESULTS
Design of the genetic code
The genotype-phenotype relationship for the population of evolving RNA enzymes was defined by a user-specified genetic code (Figure 1C). This code sought to maximize the information capacity at each genetic locus by utilizing all possible 4n combinations over four nucleotide positions (n = 4), for a total of 256 different alleles at each locus. The relationship between genotype and phenotype was not 3:1 as for the nucleotide-triplet coding of amino acids in biology, but rather a sparse code whereby a single nucleotide in the genotype region encoded either one or two nucleotides within the functional domain of the enzyme. The first, third, and fourth genotype positions each encoded two phenotype positions, thus covering one-fourth of the possible nucleotide doublets while addressing six positions within the functional domain. The second genotype position encoded only one phenotype position, providing complete sequence coverage at this position.
A different mapping rule was adopted for each genotype position (Figure 1C), again in contrast to the genetic code of biology. In order to avoid complementarity between the two genetic loci within a given enzyme, the codes for the A•B′ and A′•B loci were designed such that the same phenotype sequence was encoded by complementary genotype sequences. This strategy, however, required that the E and E′ libraries were synthesized separately. The choice of permissible nucleotides within the phenotype region was based on conservative variation of the wild-type sequence so that many of the variants were expected to retain catalytic activity (Figure 1B). However, based on knowledge of the secondary structural requirements of the enzyme (Rogers and Joyce, 2001), more than 90% of the variants were likely to be inactive.
Split-and-pool synthesis of cross-replicating RNA enzymes
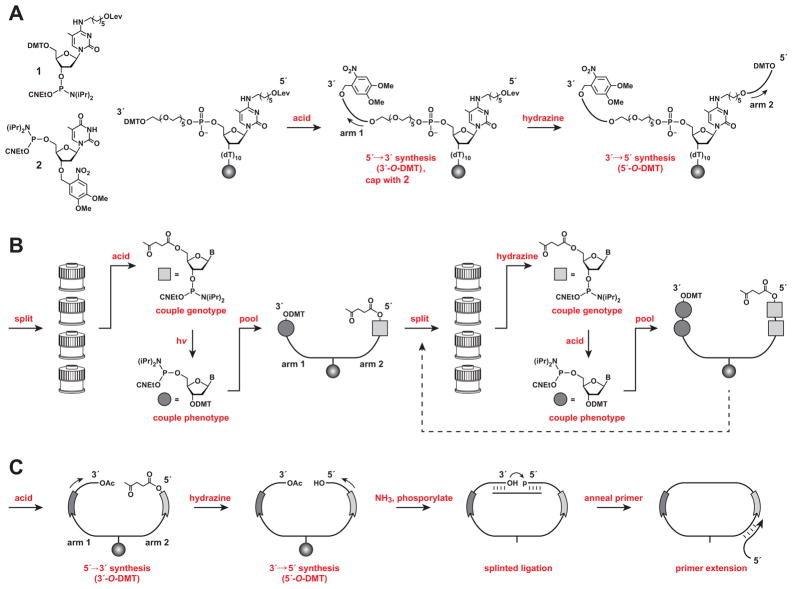
A novel split-and-pool technique (Figure 2; see also Figure S1) was used to assemble combinatorial libraries of cross-replicating RNA enzymes in accordance with the genetic code described above. Solid-phase synthesis of the corresponding DNA templates was carried out in three stages: (i) construction of a dual-armed DNA scaffold to enable tandem synthesis of the two variable regions; (ii) split-and-pool synthesis of the variable regions using orthogonally protected nucleoside phosphoramidites; and (iii) conversion of the branched molecules into linear DNA templates for in vitro transcription of the corresponding RNAs.
Figure 2.
Split-and-pool synthesis to link genotype and phenotype of cross-replicating RNA enzymes. (A) Construction of a dual-armed scaffold, linked to a solid support (sphere), followed by synthesis of the constant regions located outside the genotype and phenotype regions. (B) Split-and-pool synthesis to couple successive genotype nucleotides to arm 2 (shaded squares) and corresponding phenotype nucleotides to arm 1 (shaded circles). (C) Synthesis of the constant region of the enzyme located between the genotype and phenotype regions, followed by splinted ligation to form a closed circle, and primer extension to yield linear DNA suitable for PCR amplification and transcription. (See also Figure S1 and Table S2)
Construction of the dual-armed scaffold began by synthesizing a poly(dT) spacer, onto which was coupled a 5-Me-dC brancher phosphoramidite (1) (Figure 2A). The brancher contained a 4-N-(6-hydroxyhexyl) sidechain, protected as the levulinate (Lev), which could be selectively removed using buffered hydrazine, thus enabling the two DNA arms to be synthesized independently from a common brancher. In order to reduce steric crowding between the two arms, a polyethylene glycol spacer was coupled to the 5′ end of the brancher. Following removal of the DMT group from the spacer, a first fixed region (on arm 1) was synthesized in the 5′→3′ direction using 3′-O-DMT-protected “reverse” phosphoramidites, and capped with the photolabile 3′-O-4,5-dimethoxy-2-nitrobenzyl (nitroveratryl) phosphoramidite 2 (Wu et al., 2007; Supplemental Experimental Procedures). Then the Lev group was selectively removed and a second fixed region (on arm 2) was synthesized in the 5′→3′ direction using standard 5′-O-DMT phosphoramidites.
Next, the two variable regions were assembled, one encoded position at a time, by split-and-pool synthesis (Figure 2B). Tandem additions of a genotype nucleotide (arm 2; 3′→5′ direction) and corresponding phenotype nucleotide(s) (arm 1; 5′→3′ direction) were carried out using 5′-O-Lev (Iwai and Ohtsuka, 1988; Iwai et al., 1990) and 3′-O-DMT phosphoramidites, respectively. The Lev and DMT protecting groups were found to be highly orthogonal, provided removal of the Lev group was carried out under basic conditions (3:2 pyridine/acetic acid). However, these conditions required the use of robust nucleobase protecting groups (benzoyl for dA and dC; isobutyryl for dG) to prevent undesired deprotection and subsequent branching (Iwai and Ohtsuka, 1988).
For the first variable position, the solid support was split into four equal portions, and for each portion the 5′-O-DMT group was removed and one of four Lev-nucleotides was coupled to arm 2 (Figure 2B). Then the individual portions were briefly photolyzed to remove the 3′-O-nitroveratryl cap and the corresponding DMT-nucleotide(s) were added to arm 1 (in accordance with the genetic code; Figure 1C; see also Table S1). The split portions were reunited, mixed thoroughly, and split again. The three remaining variable positions were synthesized in a similar manner, except that the 5′ and 3′ ends of the DNA were deprotected using buffered hydrazine and acid, respectively, before adding the appropriate nucleotide(s). In addition to the 256-member library described above, a sub-library of 64 different variants, with only the first three variable genotype positions, was prepared by setting aside a portion of the pooled resin following the third split-and-pool cycle.
After synthesis of the variable regions was completed, the remainder of arm 1 was synthesized in the 5′→3′ direction using 3′-O-DMT phosphoramidites, then capped with acetic anhydride (Figure 2C). The DNA was treated for a final time with hydrazine and the remainder of arm 2 was synthesized in the 3′→5′ direction using 5′-O-DMT phosphoramidites. At this point, the entire library of dual-armed molecules was deprotected and released from the solid support using ammonium hydroxide, then purified by denaturing polyacrylamide gel electrophoresis.
Several nanomoles of purified material were obtained for each library. The purified DNA was 5′-phosphorylated and self-ligated to form circular products, which were used as templates for a primer extension reaction using DNA polymerase (Figure 2C). The resulting single-stranded DNAs were purified, cloned, and sequenced to verify that the molecules adhered to the genetic code (Table S2). The DNA then was amplified by PCR and used to transcribe the corresponding populations of either E or E′ RNA molecules. The A and A′ components were prepared from E and E′, respectively, by site-specific cleavage using E. coli M1 RNA and an external guide sequence RNA (Forster and Altman, 1990; Supplemental Experimental Procedures). This resulted in homogenous 3′ ends bearing a 2′- and 3′-hydroxyl. Synthesis of the B and B′ components was carried out by conventional synthesis, using a mixture of the four nucleoside phosphoramidites for each variable position (see Supplemental Experimental Procedures).
Sequence analysis revealed a low level of errors among the populations of E and E′ molecules, which were largely attributable to mutations during PCR amplification. Among 18 clones derived from the single-stranded DNAs, there was only one point mutation, which occurred outside the genotype and phenotype regions. Among 44 clones obtained following PCR amplification, there were five mutations that disrupted the genotype-phenotype relationship (Table S2) and six additional point mutations. Most of the former likely were due to recombination during PCR amplification (Meyerhans et al., 1990) that shuffled the genotype and phenotype regions of the otherwise identical molecules. No special precautions were taken to minimize errors during the PCR, such as prolonged extension times to reduce recombination or the use of a high-fidelity polymerase.
Cross-replication with complex mixtures
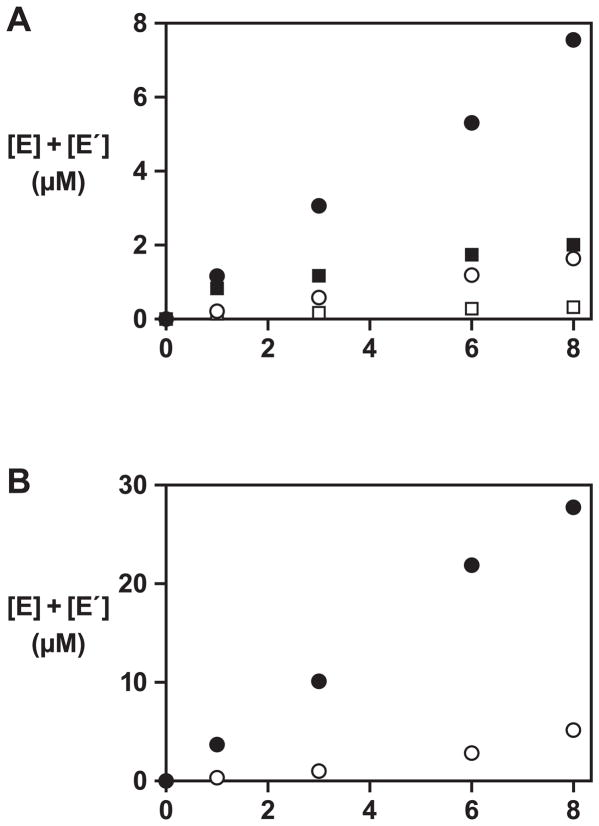
The two populations of cross-replicating enzymes had a complexity of either 4,096 (64 × 64) or 65,536 (256 × 256) different combinations. These populations were tested separately in an amplification reaction employing 5 μM total concentration of E and E′ and 50 μM total concentration of each of the four substrates (A, A′, B, and B′). The reactions were carried out in the presence of 40 mM MgCl2 at pH 8.5 and 42 °C. A modest amount of product (~8% yield) was formed after 8 h with the population of 4,096 different, whereas a control reaction containing only the four substrates did not yield a significant amount of product (Figure 3A). Only a small amount of product (~2% yield) was obtained in the reaction with the more diverse population of 65,536 different enzymes. Prolonged incubation times or alternative reaction conditions failed to increase the yield with the more diverse population. This may not be surprising considering that >90% of the molecules lacked catalytic activity, but it demonstrates that the active enzymes could not overcome the larger number of inactive molecules. Therefore, it was concluded that the library of 65,536 different enzymes exceeded the complexity that could be supported by the genetic system implementing this particular genetic code. Subsequent evolution experiments were initiated using the less diverse population of cross-replicating enzymes.
Figure 3.
Cross-replication reactions employing the starting population of enzymes. (A) Amplification of a population of enzymes comprised of either 4,096 (circles) or 65,536 (squares) different variants, initiated with either 5 μM total concentration (black) or no starting amount (white) of both E and E′. The reaction mixture contained 50 μM total concentration of each of the four substrates (A, A′, B, and B′), 40 mM MgCl2, and 50 mM EPPS (pH 8.5), and was incubated at 42 °C. (B) Amplification of the enriched population of enzymes and substrates obtained after the 10th round of growth and dilution, initiated with either 5 μM total concentration (black) or no starting amount (white) of both E and E′. Reaction conditions are as described in (A).
Self-sustained evolution of RNA
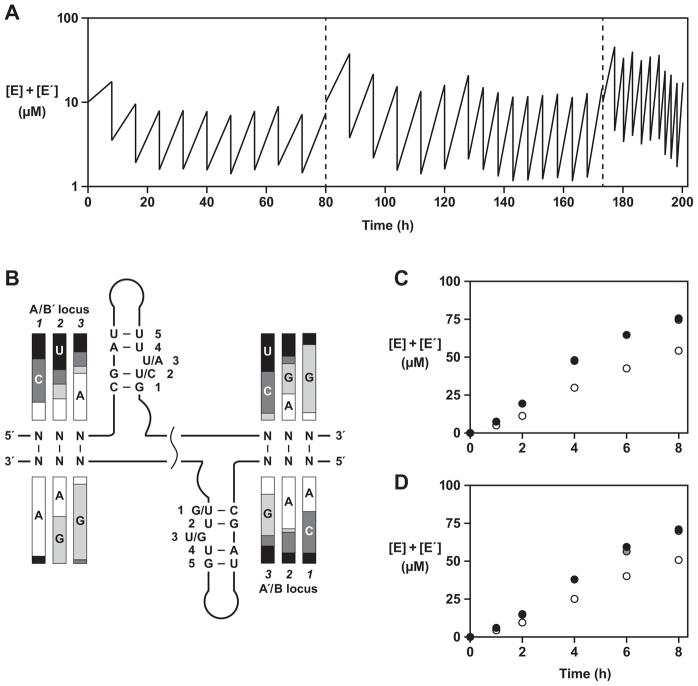
Cross-replication could be sustained indefinitely through a serial transfer process involving repeated transfer of a portion of a completed reaction mixture to a new vessel containing fresh reaction components. Ten successive reactions were carried out in this fashion using the population of 4,096 different cross-replicating enzymes (Figure 4A). The duration of each reaction was 8 h, and 20% of the material was transferred from one reaction mixture to the next. The initial reaction mixture was seeded with 5 μM total concentration of both E and E′ and all of the reaction mixtures contained 50 μM total concentration of each of the four substrates. No additional enzymes were added to subsequent reaction mixtures other than those that were carried over in the transfer. Cross-replication was maintained for a total of 80 h incubation, resulting in ~107-fold overall amplification.
Figure 4.
Self-sustained evolution of RNA enzymes. (A) Serial transfer procedure involving 35 successive rounds of growth and dilution. After the 10th round (left dashed line), an enriched set of substrates was prepared from the population of enzyme molecules and used in subsequent rounds. After the 25th round (right dashed line), the same enriched set of A and A′ substrates was used, together with B59 and B′213. (B) Sequence variation among 47 clones isolated after the 25th round (see also Table S3). The relative proportions of the four nucleotides at each genotype position are indicated by shaded bars. The most abundant nucleotides at each phenotype position are shown. (C, D) Cross-replication reactions employing: 5 μM each of the population of E and E′ molecules obtained after the 25th round; 50 μM each of the enriched set of A and A′ molecules obtained after the 25th round; either (C) 50 μM B′213 or (D) 50 μM B′245; and either 50 μM B27 (white circles), 50 μM B51 (grey circles), or 50 μM B59 (black circles). (See also Figure S2)
The E and E′ products from the final reaction mixture were isolated by polyacrylamide gel electrophoresis, reverse transcribed, and amplified by PCR. The resulting DNA was used to prepare an enriched library of substrates (A, A′, B, and B′), derived from the evolved population of enzymes (see Supplemental Experimental Procedures). This was done to reduce the diversity of the substrates to match the reduced diversity of the more selectively advantageous enzymes. Cross-replication using the evolved enzymes and enriched library of substrates was much more efficient, with ~30% product formed after 8 h, compared to ~8% for the naive population (Figure 3B).
The enriched library was used to carry out a second, more aggressive serial transfer experiment in which 10% of the material was transferred from one reaction mixture to the next (Figure 4A). Again, each of the four substrates was present at a total concentration of 50 μM. The yield of products declined somewhat during the first three reactions, then began to rise. After the sixth transfer, the incubation time was reduced from 8 to 5 h to increase the selection stringency imposed on the evolving population. A total of 15 successive reactions were carried out over 93 h, resulting in ~1015-fold overall amplification.
The products from the final reaction mixture were isolated, cloned, and sequenced (Table S3). This analysis revealed that no single pair of cross-replicating enzymes had grown to dominate the population. Of 47 clones that were sequenced, only three occurred more than twice and 34 were unique. Interestingly, there was only modest complementarity between the consensus sequences at each of the two genetic loci (Figure 4B). At the A•B′ locus, for example, only a single E clone exhibited perfect complementarity to the B′ portion of E′. This suggests that the fidelity of replication is low and that perfect complementarity between the enzymes and their substrates is not required, or perhaps not even advantageous, for survival during the course of evolution. Also, as a consequence of the additional PCR amplification steps used to prepare the enriched library of substrates, about one-third of the clones contained mutations that disrupted the genotype-phenotype relationship (Table S3), and there were 17 additional point mutations outside the genotype and phenotype regions (error rate = 0.42% per nucleotide position).
When the individual components (A, A′, B, and B′) of the cloned enzymes were considered separately, it became clear that the variable regions within the B and B′ components were more enriched compared to those within the A and A′ components (Table S3). For example, two B′ variants (B′213 and B′245) accounted for ~80% of the E′ clones, whereas no single A or A′ variant occurred more than three times. This observation suggested that the system had become enriched with a few “universal” B and B′ substrates, which were utilized by various enzymes during cross-replication, despite the imperfect complementarity.
In order to test this hypothesis, the five most abundant B and B′ substrates were synthesized individually and all possible combinations were examined independently in cross-replication reactions employing the evolved population of E and E′ together with the enriched set of A and A′ molecules. All cross-replication reactions exhibited exponential amplification, reaching a maximum extent of >50% during the 8 h incubation (Figure 4C, D). However, when a pair of the less abundant B and B′ substrates were tested similarly, much lower product formation was observed (<15%) (Figure S2). These data verified that the enriched B and B′ substrates were utilized by a variety of cross-replicating enzymes within the population, and suggested that the enriched B and B′ substrates were preferred in this regard.
A final serial transfer experiment was carried out using only the most efficient B and B′ variants (B59 and B′213), together with the A, A′, E, and E′ molecules that had emerged from the prior evolution experiment (Figure 4A). Ten successive reactions were carried out over 26 h, resulting in ~1010-fold overall amplification. The products from the final reaction mixture were isolated by polyacrylamide gel electrophoresis, cloned, and sequenced (Figure 5A). About half of the variants occurred multiple times among 26 clones that were sequenced, indicating that the population had undergone further selective enrichment. The single most abundant E variant was E(A65-B59), which occurred three times. This suggests either that B′213 is an excellent substrate for E(A65-B59), or that A65 is an excellent substrate for E′ containing the B′213 component, despite poor complementarity within the genetic region in either case. Complementarity of the A•B′ locus was generally poor, with the majority of genetic alleles containing at least two mismatches. However, complementarity at the B•A′ locus was much higher, with all but two clones having no more than a single mismatch at this locus.
Figure 5.
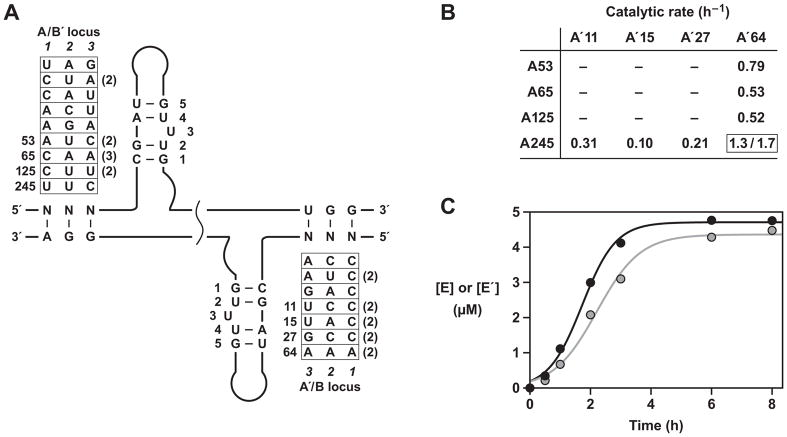
Properties of the final evolved RNA enzymes. (A) Sequence variation among 26 clones isolated after the 35th round of self-sustained evolution. All E or E′ molecules contained component B59 or B′213, respectively, and contained various A or A′ components as indicated. The A and A′ components that were tested individually are numbered. The numbers in parentheses indicate duplicate clones. The most abundant nucleotides at each phenotype position are shown. (B) Reactions employing 5 μM each of B59, B′213, and various combinations of the most abundant A and A′ molecules, together with 0.1 μM each of the corresponding E and E′ molecules (see also Figure S3). There was no detectable activity when neither A245 nor A′64 was used, only linear amplification when either A245 or A′64 was used (linear rate shown for production of either E′ or E, respectively), and exponential amplification when both A245 or A′64 were used (exponential rate shown for production of E′/E). (C) Cross-replication reaction employing 0.1 μM each of E(A245-B59) and E′(A′64-B′213) and 5 μM each of A245, A′64, B59, and B′213, resulting in exponential amplification of both E (black) and E′ (grey).
Properties of individual evolved enzymes
Several of the most abundant evolved cross-replicating enzymes were prepared and tested individually in cross-replication reactions, together with their corresponding substrates. The reaction mixtures contained 5 μM each of the four substrates and a starting concentration of 0.1 μM each of E and E′. Surprisingly, the only pair of enzymes that underwent exponential amplification was E(A245-B59) in combination with E′(A′64-B′213) (Figure 5B, C). Only linear amplification was observed when either of these enzymes was paired with a different cross-replicating partner. Reaction mixtures that did not contain either E(A245-B59) or E′(A′64-B′213) did not produce any detectable product. However, A′ and A molecules derived from the enriched but inactive enzymes were excellent substrates for E(A245-B59) and E′(A′64-B′213), respectively (Figure S3). These data show that the catalytic activity of the two highly reactive cross-replicating enzymes is sufficient to sustain the growth of many inactive enzymes, the latter surviving as molecular parasites by exploiting the modest fidelity of the genetic system.
DISCUSSION
The synthetic genetic system based on cross-replicating RNA enzymes entails two genetic loci of 6–7 nucleotides each. In principle, this system could support ~104–105 different alleles at each locus, for a combinatorial complexity of 108–1010 different variants. Such diversity would be much greater than what is typically accessed through combinatorial chemistry, and likely would enable the emergence of novel RNA-based functions during the course of self-sustained evolution. Unlocking this high diversity requires a means for generating complex libraries of replicating RNAs that enforce the genotype-phenotype relationship on a molecule-by-molecule basis. It also requires establishing a genetic code that operates with sufficient fidelity to maintain heritable information throughout the course of evolution.
The synthetic challenge of accessing large numbers of genotype-phenotype pairs has now been met through a split-and-pool approach that relates two different sequence regions within the same nucleic acid molecule. One of these regions plays a genetic role and the other forms part of the catalytic center of the enzyme. The encoded phenotype region could instead be appended to the catalytic center, for example, as part of an aptamer domain that causes replication to be contingent upon the aptamer binding its cognate ligand (Lam and Joyce, 2009).
The present study employed populations of either 4,096 or 65,536 different cross-replicating RNA enzymes, which are the most complex non-biological genetic systems ever constructed. The same synthetic approach could be used to prepare much more complex populations, with ten cycles of split-and-pool synthesis likely to be readily achievable. However, such complexity would far exceed the capacity of the system to propagate genetic information, which already was the case here for the 65,536-member population having all possible four-nucleotide combinations at each genetic locus.
There are other potential applications of the synthetic method used to prepare nucleic acid molecules containing two related regions of variable sequence. For example, one could synthesize a population of random-sequence, fully paired helices or generate a complex mixture of molecules containing various matched tertiary structural elements. One could think more generally of preparing sets of nucleic acid molecules containing two addressable sequence elements that are related on a molecule-by-molecule basis through some arbitrary coding scheme. For these applications, it should be possible to achieve a complexity of >106 distinct related sequence elements in a single synthesis.
With regard to the population of cross-replicating RNA enzymes, the informational challenge is more formidable than the synthetic one because it requires discrimination among numerous genotypes that must operate in parallel, all drawing upon a common set of oligonucleotide substrates. A mismatch between one of the genotype regions of a parent molecule and a corresponding oligonucleotide component of a progeny molecule results in a “mutation”. Occasional mutations are desirable because they maintain diversity in the evolving population, but if mutations are too frequent there is the risk of loss of genetic information. Generally, if the number of replicatable error copies of an advantageous molecule exceeds the number of accurate copies, then the fittest molecules cannot be enriched by selection (Eigen, 1971).
The populations of cross-replicating enzymes employed in this study were based on a genetic code that allowed any of the four nucleotides to occupy each of the variable positions within the two genetic loci. The identity of each genotype nucleotide determined a component of the corresponding phenotype. This is in contrast to the genetic code of biology, where 61 different nucleotide triplets (excluding stop codons) encode only 20 different amino acids. The degeneracy of the biological code, especially at the third (wobble) codon position, causes the most common mismatches between codon and anticodon to have no effect on phenotype. A complete and non-degenerate code as was employed here is maximally susceptible to mismatches, thus providing a stringent test of the maximum coding capacity of the system.
The high potential for mismatches between parent molecules and the substrate components of their progeny led to the emergence of “universal” B and B′ components that function well in amplification reactions with a diverse set of A and A′ components (Figure 5B, C). Only a subset of the evolved RNAs were found to have robust catalytic activity, and these catalysts sustained the population as a whole. The other evolved RNAs survived as parasites, lacking intrinsic catalytic activity, but being perpetuated by the active catalysts.
The most abundant active catalysts contained either the wild-type form of the catalytic center (A245) or a permissible variant (A′64) that replaces a U•G pair by a U•A pair (Figure 5A). The most abundant parasites contained disruptive mutations within the catalytic center that introduced either one (in A53, A65, and A′27) or two (in A′11 and A′15) base mismatches or that converted a critical G•U pair to a G•C pair (in A65 and A125). The latter change was previously shown to disrupt catalytic activity without changing the secondary structure of the catalytic center (Lincoln and Joyce, 2009). The distribution of selected phenotypes is somewhat different for the E and E′ molecules (Figure 4B), which likely reflects the asymmetry of the genetic code for the two enzymes.
It would be possible to design an alternative genetic code that is less susceptible to mismatches between parent molecules and progeny components, thus disfavoring the emergence of universal B and B′ components and the survival of parasites. For example, a rule could be adopted that all possible genotypes differ by at least two nucleotides. As in biology, more than one genotype nucleotide could be used to encode each component of the corresponding phenotype. A sparse code could be employed such that every nucleotide within a given codon is distinct from the corresponding nucleotide within all of the other codons. The genetic code might also be optimized with regard to the reaction conditions, including substrate concentration, Mg2+ concentration, pH, and temperature, all of which can affect the susceptibility to mismatches between the parent molecules and progeny components.
There is ongoing debate regarding the extent to which the genetic code in biology was determined by chemical features of the interaction between codons (or anticodons) and their corresponding amino acids (the stereochemical hypothesis) or was the result of chance assignments that were locked in place by the many interdependencies of the translation apparatus (the frozen accident hypothesis; Crick, 1968; Copley et al., 2005; Woese and Goldenfeld, 2009). For synthetic genetic systems that are prepared by split-and-pool synthesis, the code is neither frozen nor an accident. New codes can be designed and tested based on cycles of hypothesis and experimentation. The best codes will be those that strike the right balance between maximizing information capacity and maintaining sufficient fidelity to ensure the enrichment of advantageous traits.
SIGNIFICANCE
A central aim of synthetic biology is to understand the principles and processes of biology by constructing alternative forms of biological systems. One of the most fundamental processes of biology is genetic inheritance, which provides the basis for Darwinian evolution. A synthetic evolving system has been constructed based on populations of cross-replicating RNA enzymes that compete for limited resources. Heretofore, each member of the population and its component substrates had to be synthesized individually, which precluded the investigation of large, complex populations and the implementation of alternative genetic codes for relating genotype and phenotype. A novel strategy for encoded combinatorial chemistry now makes it possible to construct highly complex populations of replicating enzymes, employing a user-specified genetic code that links genotype and phenotype for each molecule in the population. Such a population was made to undergo self-sustained Darwinian evolution, resulting in the selective enrichment of the most advantageous variants. These included both highly active enzymes and molecular parasites with low catalytic activity. This outcome demonstrates the trade-off between information capacity and fidelity of a genetic code, and suggests how cycles of design and experimental testing can be used to refine the properties of a code.
EXPERIMENTAL PROCEDURES
Split-and-pool synthesis of cross-replicating RNA enzymes
Solid-phase DNA synthesis was carried out using an Applied Biosystems Expedite 8900 automated DNA/RNA synthesizer (Foster City, CA). All DNA synthesis reagents and nucleoside phosphoramidites were purchased from Glen Research (Sterling, VA), except the 5′-O-Lev phosphoramidites and reverse 3′-O-DMT-deoxyguanosine phosphoramidite, which were from ChemGenes (Wilmington, MA). Synthesis of phosphoramidite 2 is described in the Supplemental Experimental Procedures. For each split-and-pool synthesis, two 1,000-Å pore size dT columns (1 μmol scale) were used. DNA sequences and phosphoramidite coupling protocols are described in the Supplemental Experimental Procedures and Figure S1.
The Lev protecting group was removed from the support-bound material, still within the synthesis column, which was dried under vacuum and placed under an argon atmosphere. Using two syringes, one at each end of the column, 0.7 ml of hydrazine hydrate solution (0.25 M in 3:2 pyridine/acetic acid) was forced back-and-forth across the resin for 10 min. The resin then was washed with 10 ml of 3:2 pyridine/acetic acid solution and 10 ml of acetonitrile, and dried under vacuum.
Split-and-pool synthesis began with the first variable position, employing the support-bound material as a slurry in acetonitrile, which was divided into four equal portions and transferred into separate synthesis columns. Each portion was dried under vacuum and the appropriate Lev nucleotide was coupled to arm 2 (Figure S1). Then each portion was transferred to a small glass vial and suspended in 1 ml of 10:1 acetonitrile/water. The slurries were photolyzed (350 nm) at 4 °C for 30 min, with gentle agitation every 5 min. Stir bars were avoided to prevent damage to the controlled pore glass support. After photolysis, the solutions were decanted, the resin was washed twice with 2 ml of acetonitrile, and the material was returned to the four separate synthesis columns, where the appropriate DMT nucleotide was coupled to arm 1. The separate portions of resin then were pooled in a small glass vial containing 2 ml acetonitrile and gently mixed for 10 min. Subsequent rounds of split-and-pool synthesis were carried out similarly, with the 3′-O-DMT group on arm 1 removed using the synthesizer’s standard detritylation cycle.
After synthesis of the variable regions was complete, the remaining constant region of arm 1 was synthesized using standard methods (Figure S1). The completed arm 1 was capped using acetic anhydride, the 5′-O-Lev group on arm 2 was removed using hydrazine (as above), the remaining constant region on arm 2 was synthesized, and the entire dual-armed molecule was deprotected and released from the solid support using 28% aqueous ammonium hydroxide, incubating at 55 °C for 16 h. Procedures for circularization of the dual-armed molecules, formation of linear DNA templates, and transcription of the corresponding RNAs are described in the Supplemental Experimental Procedures.
Serial transfer experiments
Exponential amplification of the RNA enzymes was carried out in a reaction mixture containing 50 μM each of the A, A′, B, and B′ substrates, 40 mM MgCl2, and 50 mM EPPS (pH 8.5), which was incubated at 42 °C. The first reaction mixture was seeded with 5 μM each of E and E′, and all subsequent reaction mixtures contained only the enzymes that were carried over in the transfer. Reactions were initiated by mixing equal volumes of two solutions, one containing A, A′, E and E′ and the other containing B, B′, MgCl2, and EPPS.
Supplementary Material
Acknowledgments
This work was supported by grant NNX07AJ23G from NASA, grant R01GM065130 from the NIH, and by The Skaggs Institute for Chemical Biology at The Scripps Research Institute. J.T.S. was supported by Ruth L. Kirschstein National Research Service Award No. F32 GM101741 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brenner SA, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nature Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- Copley SD, Smith E, Morowitz HJ. A mechanism for the association of amino acids with their codons and the origin of the genetic code. Proc Natl Acad Sci USA. 2005;102:4442–4447. doi: 10.1073/pnas.0501049102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwiss. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Forster AC, Altman S. External guide sequence for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Furka Á, Sebestyén F, Asgedom M, Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Peptide Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin DR, Harbury PB DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2004;2:1022–1030. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghten RA, et al. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Iwai S, Ohtsuka E. 5′-Levulinyl and 2′-tetrahydrofuranyl protection for the synthesis of oligoribonucleotides by the phosphoramidite approach. Nucleic Acids Res. 1988;16:9443–9456. doi: 10.1093/nar/16.20.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S, Sasaki T, Ohtsuka E. Large scale synthesis of oligoribonucleotides on a solid support: synthesis of a catalytic RNA duplex. Tetrahedron. 1990;46:6673–6688. [Google Scholar]
- Kim D-E, Joyce GF. Cross-catalytic replication of an RNA ligase ribozyme. Chem Biol. 2004;11:1505–1512. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Lam BJ, Joyce GF. Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nature Biotechnol. 2009;27:288–292. doi: 10.1038/nbt.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nature Biotechnol. 2004;22:568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- Meyerhans A, Vartanian J-P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;22:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc Natl Acad Sci USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Joyce GF. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LA, Ellman JA. Synthesis and applications of small molecule libraries. Chem Rev. 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- Woese CR, Goldenfeld N. How the microbial world saved evolution from the Scylla of molecular biology and the Charybdis of the modern synthesis. Microbiol Mol Biol Rev. 2009;73:14–21. doi: 10.1128/MMBR.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. 3′-O-modified nucleosides as reversible terminators for pyrosequencing. Proc Natl Acad Sci USA. 2007;104:16462–16467. doi: 10.1073/pnas.0707495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.