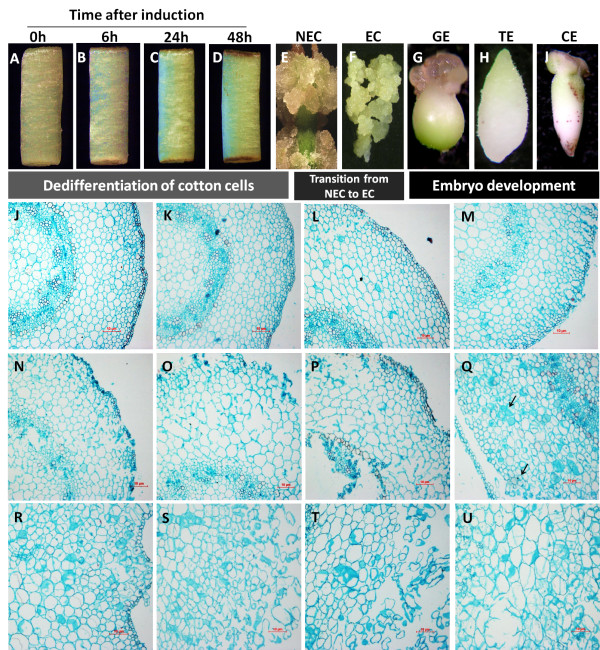
Figure 1.
Schematic representation and histological observation of different time points/stages during somatic embryogenesis used for RNA-Seq analysis. Initial hypocotyl explants (A) used as control. Initial cellular dedifferentiation was sampled from explants after induction for 6 h, 24 h, and 48 h (B-D). Typical nonembryogenic calli (NECs) were sampled at 40 d of culture time (E) when the calluses were loosening and abundant, embryogenic calluses (ECs) were sampled after the first subculture when compact primary embryogenic clumps (F) were first identified. Different stages of somatic embryos [globular embryos (GEs), (G), torpedo embryos (TEs), (H), and cotyledon embryos (CEs), (I)] were sampled after synchronization control of somatic embryogenesis by suspension culture. Somatic embryogenesis in cotton passes through three different processes: dedifferentiation of cotton somatic cells, transition from NECs to ECs, and development of Somatic embryos. Histological analysis was made at 0 h, 3 h, 6 h, 12 h, 24 h, 48 h, 72 h, 7 d, 10 d, 15 d, 25 d, and 40 d after induction (J-U).